Gene of encoded cytosine deaminease for preventing and controlling crop disease
A cytosine deaminase and plant disease control technology, applied in the direction of introducing foreign genetic material, application, animal repellent, etc. by using vectors, can solve problems such as aggravating environmental pollution, drug residues, and increasing agricultural costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Construction of Xanthomonas mutant library of cabbage black rot
[0027] Tn5gusA5 (Sharma, 1991) was introduced into Xcc wild-type strain 8004 using the shuttle plasmid pLAFR1 between E. Namycin) resistance marker screening Xcc::Tn5gusA5 insertion mutants, combined with Xcc whole genome sequence (genome sequence number NC 007086) and TAIL-PCR (Thermal Asymetric Interlaced-PCR) (Yao-Guang Liu, 1995) technology, to determine The insertion position of Tn5gusA5 on the genome, and the insertion position was verified by PCR. By investigating the phenotypic changes such as pathogenicity, extracellular enzyme activity, and extracellular polysaccharide synthesis of mutants, the pathogenicity-related genes are screened (Tang et.al, 2005). The present invention relates to one of the new pathogenicity-related genes- XC1949 gene (cytidine deaminase gene), its insertion mutant number is 118H11, the screening process can be found in the reference (Tang et.al, 2005).
Embodiment 2
[0028] Example 2 Cloning and sequence determination of XC1949 gene (cytosine deaminase gene)
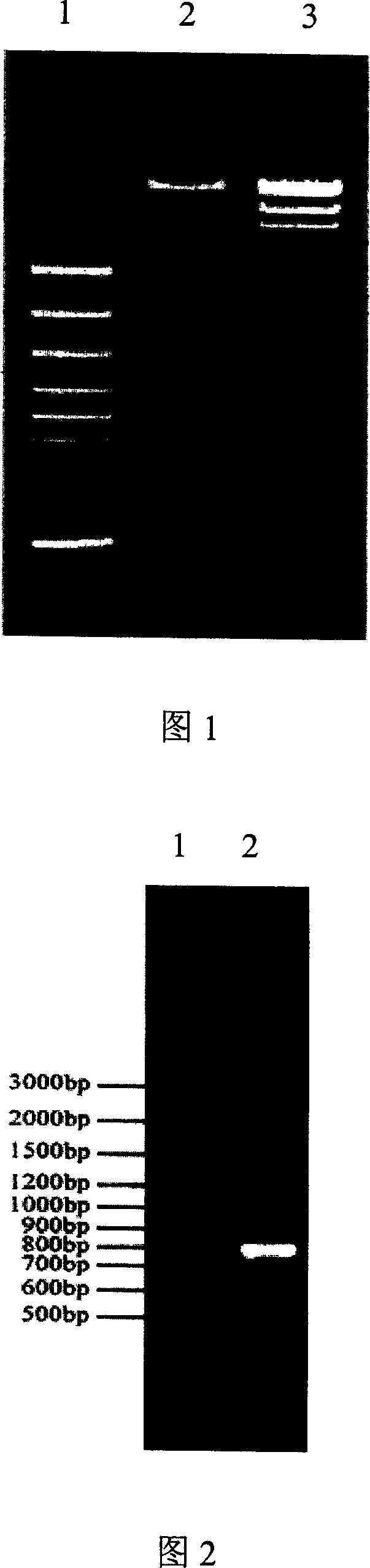
[0029] According to the gene sequence of XC1949 (see sequence table SEQ ID No.1, base 1 to base 927), design primers (upstream primer GGGAATTCATCGCGGCGTGTTTTTGACCCC and downstream primer GGTCTAGACTCGGCGAGCACGTTCAACTGC; PCR conditions 95°C for 3min; 95°C for 30sec, 60°C for 30sec , 72°C for 1min, 30 cycles; 72°C for 5min)), using the total DNA of Xanthomonas cabbage black rot strain 8004 as a template, the full-length sequence of the gene was amplified by PCR, and cloned into the cloning vector pGEM3Zf In (+), the DNA nucleotide sequence (SEQ ID No.1) was determined on an ABI 377 DNA automatic sequencer by the dideoxynucleotide method. The correct XC1949 gene sequence verified by sequencing was cloned into the shuttle vector pLAFRJ, and the recombinant plasmid pCXC1949 containing the gene was obtained. The plasmid was digested with EcoRI and XbaI, and besides a 22kb vector band, ther...
Embodiment 3
[0030] Example 3 Verification of XC1949 Gene Insertion Mutant 118H11
[0031] The insertion mutant 118H11 was sequenced and located by TAIL-PCR, and it was found that it was inserted into the XC1949 gene. Use a primer on the gus side of transposon Tn5 (TTGTAACGCGCTTTCCCACCAACGC) and a primer (5'-GGGCAAGAGCGAAAAGCTGGACG-3') of the upstream gene XC1948 to perform PCR verification (95°C 3min; 95°C 30sec, 60°C 30sec, 72°C 45sec , 30 cycles; 72° C. for 5 min), the expected size of the product is 788 bp (see Figure 2).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com