Identification of transglutaminase substrates and uses therefor
a transglutaminase and substrate technology, applied in the direction of transferases, peptide sources, plasma/lacto globulins, etc., can solve the problems of difficult finding common substrate motifs, for example transglutaminase substrate motifs, and limited identification of synthetic peptides as enzyme substrates, so as to achieve optimal enzyme specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
elopment
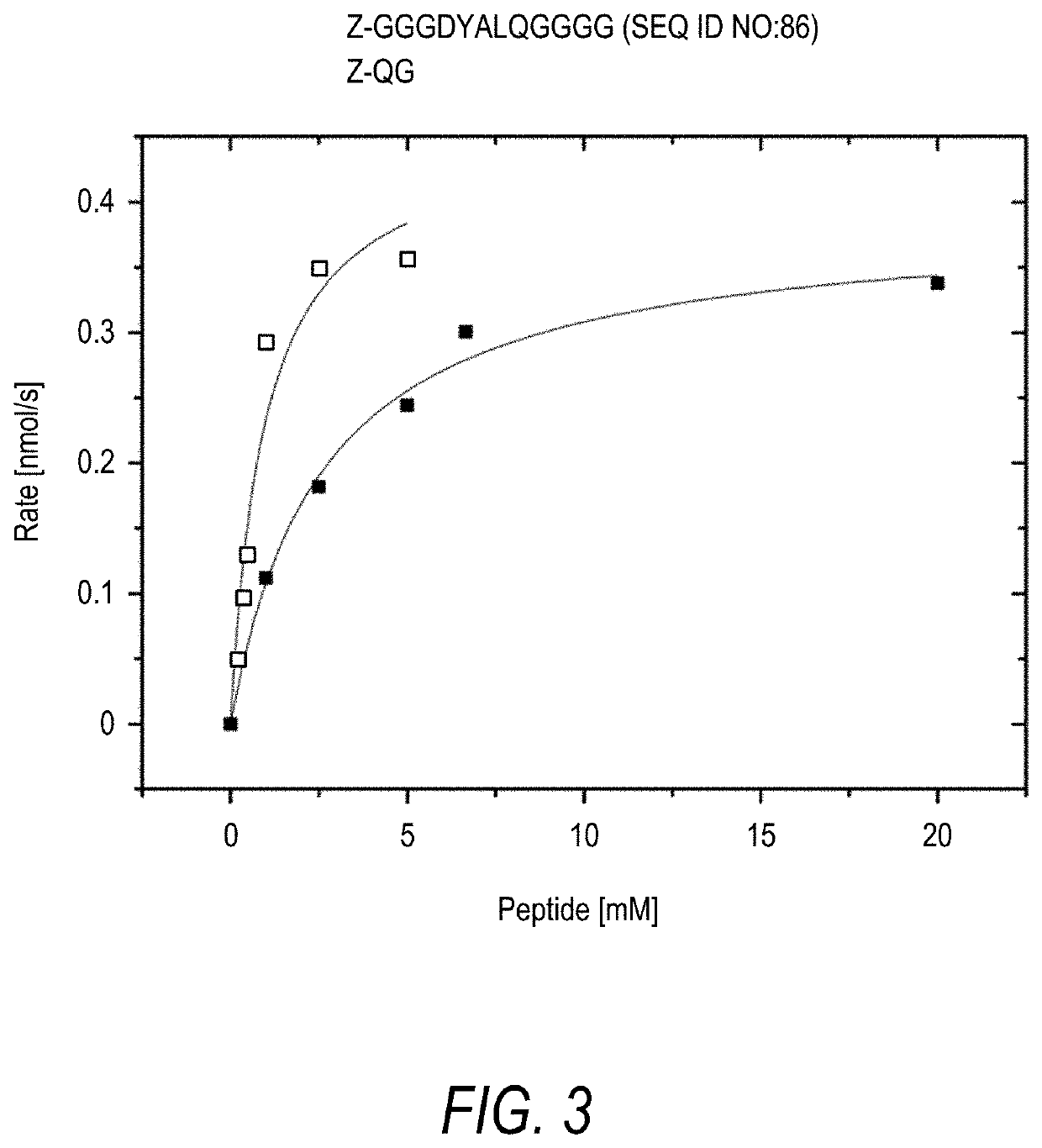
[0232]To test MTG (Zedira GmbH) specificity for Gln-substrate, N-(Biotinyl)cadaverine (Zedira GmbH) was used as a substitute for a Lys-substrate to biotinylate Gln-peptides on a peptide array synthesized using maskless light-directed peptide array synthesis. Similarly, to test MTG specificity for Lys-substrate, Z-Gln-Gly-CAD-Biotin (Zedira GmbH) was used as a substitute for a Gln-substrate to biotinylate Lys-peptides. Z-Gln-Gly-CAD-Biotin is a glutamine donor substrate for transglutaminases having the following formula:
[0233]
[0234]After treatment with MTG in the presence of one of the biotinylated substrates, arrays were washed, stained with Cy5-streptavidin to label biotin moieties, and scanned at 635 nm to measure signal intensity at the peptide areas. Signal intensity corresponding to the efficiency of MTG reaction was used to determine specificity of different peptide sequences.
example 2
ditions
[0235]In general, there were two issues to consider in optimizing array performance of the enzymatic assay originally developed under solution conditions. A first issue was a challenge of low signal generation that could be caused by the inability of the enzyme to recognize peptides bound to the surface, low peptide concentration, insufficient quality of peptide synthesis, or by surface effect on enzyme stability and reactivity. A second issue was a challenge of high background generation that may be a result of non-specific binding of the enzyme and / or a substrate to an array surface or non-specific labeling driven by side reactions on the array.
[0236]To find conditions for MTG assay on the array, the effects of various parameters of the reaction on signal and background generation were analyzed. These factors included MTG and substrate concentrations, reaction buffer composition, and incubation time and temperature. In order to minimize background, a non-protein blocking so...
example 3
ficity for Gln-Peptides
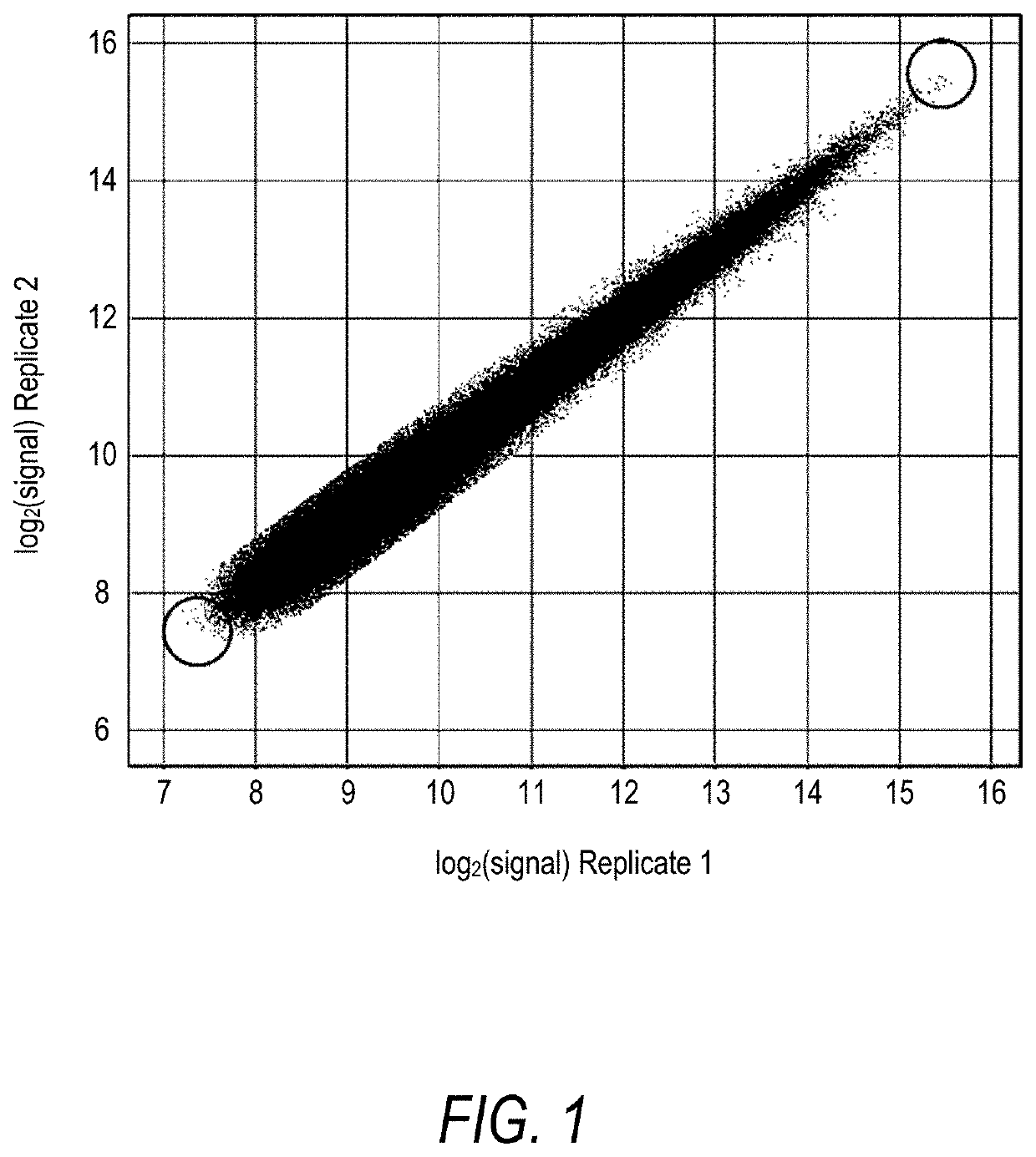
[0238]A 5-mer peptide array was incubated in the presence of MTG and biotinylated amine donor N-(Biotinyl)cadaverine substrate under conditions described in Example 2. Signal distribution and correlation between two replicates was plotted (FIG. 1).
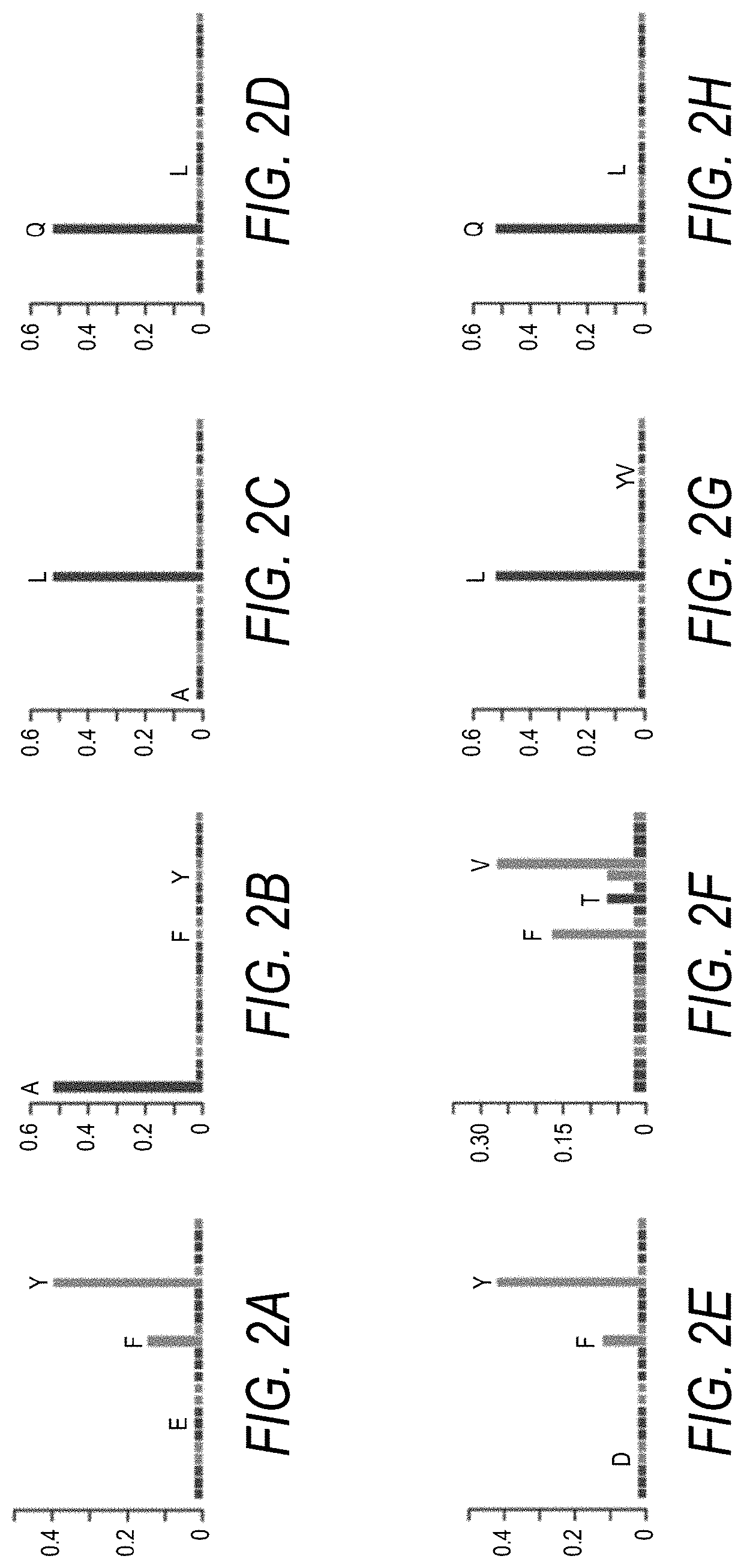
[0239]With reference to FIG. 1, correlation in signal intensity between replicates showed high reproducibility of the data and allowed identification of peptides with the highest labeling efficiency. Sequences and corresponding signal intensity of 25 peptides with the highest labeling efficiency in an array MTG assay with biotinylated amine-donor substrate discovered in this study are shown in Table 2. In concordance with MTG specificity, all peptides contained a Gln (Q) residue. Gln occupied exclusively the fifth position in all selected sequences. Because the 5-mer peptides synthesized on the array were flanked by G:S linkers for which a 3:1 mixture of Gly and Ser amino acid precursors was used, Gln may be followed...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| molecular weights | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com