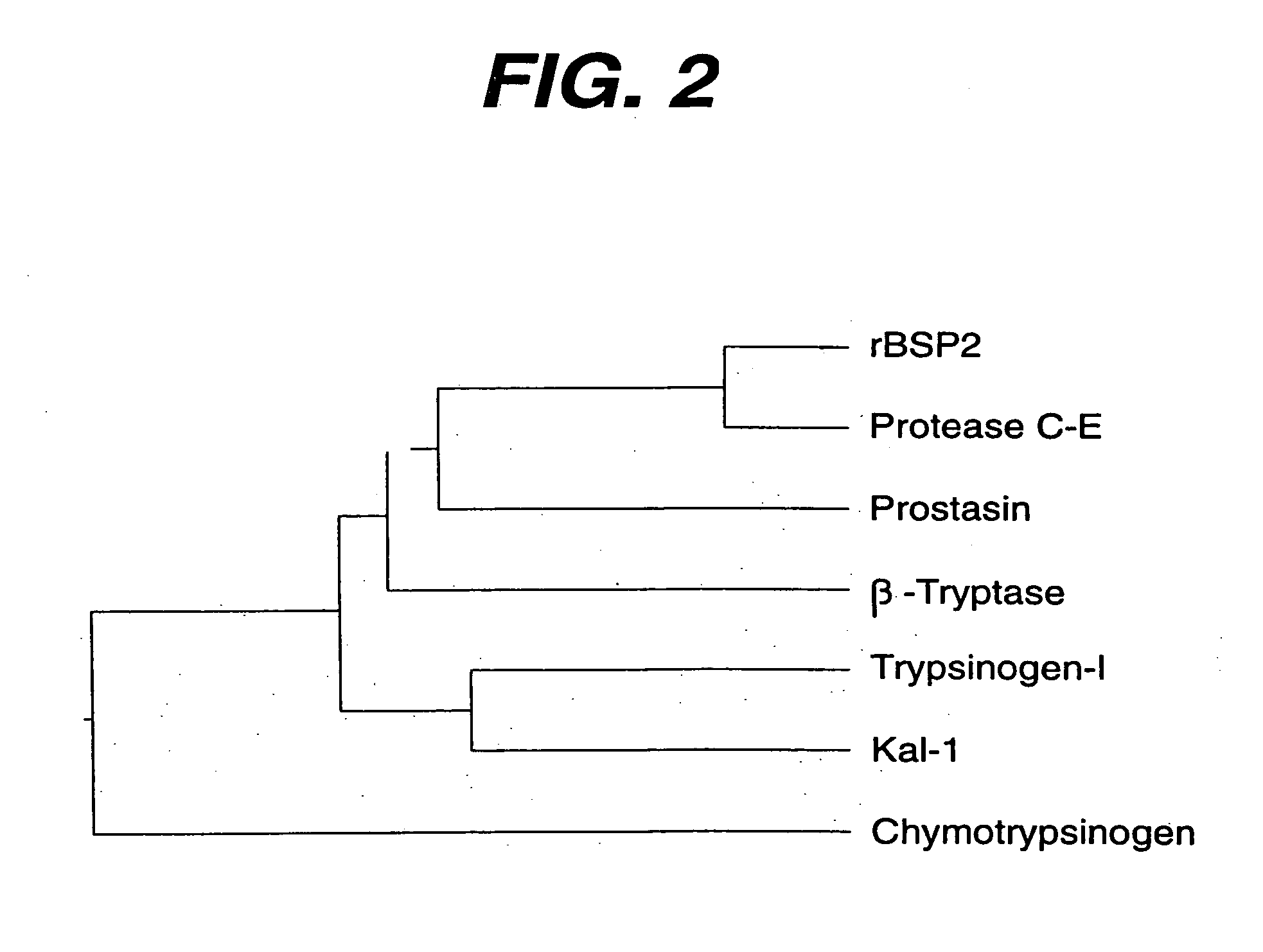
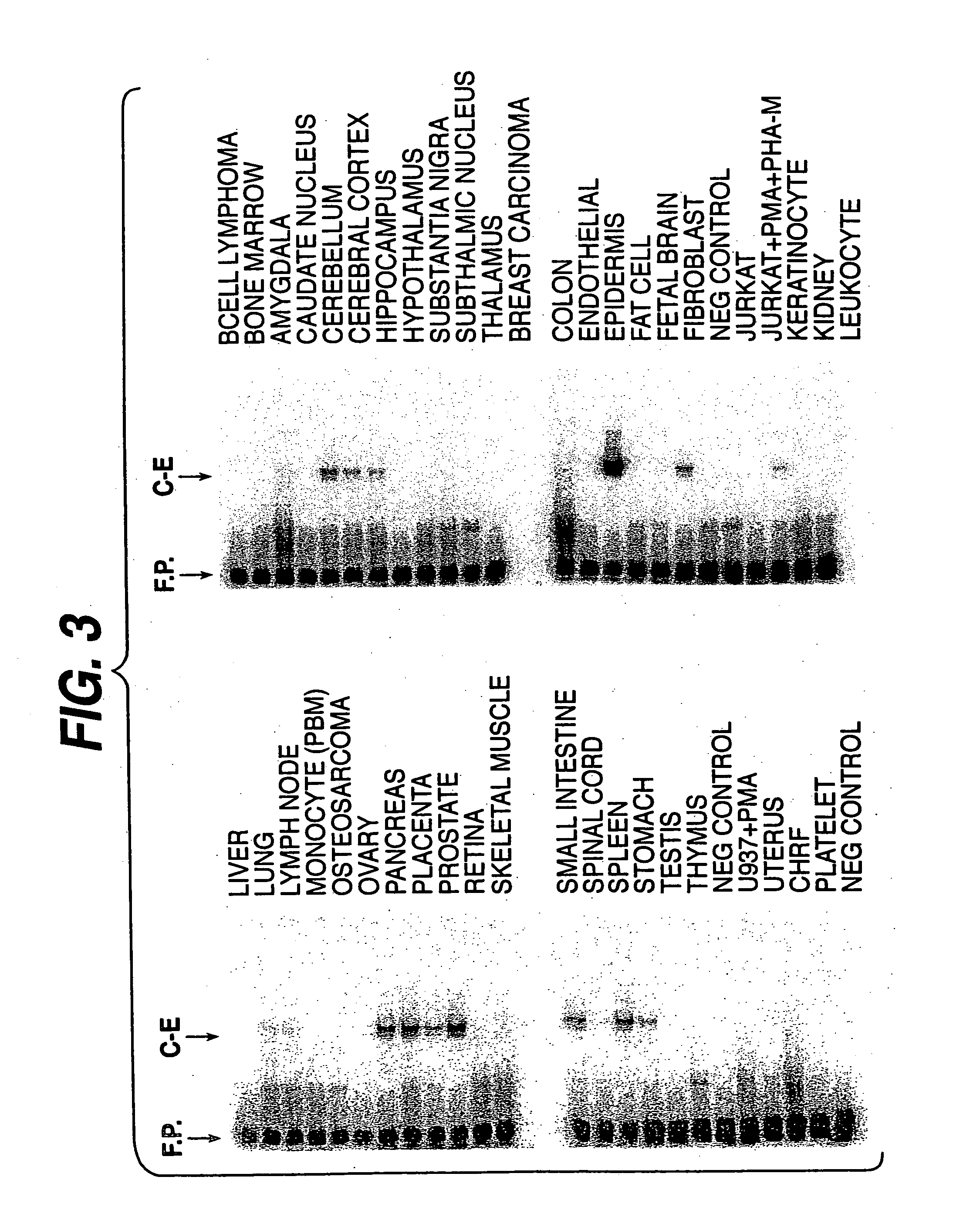
DNA encoding the human serine protease C-E
a technology of serine protease and dna encoding, which is applied in the direction of hydrolases, peptide/protein ingredients, fungi, etc., can solve the problems of limited use of some proteases and reduced efficiency when used in a non-natural environmen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Plasmid Manipulations:
[0128] All molecular biological methods were in accordance with those previously described (Maniatis et al. (1989). 1-1626). Oligonucleotides were purchased from Ransom Hill Biosciences (Ransom Hill, Calif.) and all restriction endonucleases and other DNA modifying enzymes were from New England Biolabs (Beverly, Mass.) unless otherwise specified. The protease C-E expression construct was made in the baculovirus expression vector pFastBac1 (Life Technologies, Gaithersberg, Md.) as described below. All construct manipulations were confirmed by dye terminator cycle sequencing using Allied Biosystems 377 fluorescent sequencers (Perkin Elmer, Foster City, Calif.).
Acquisition of Protease C-E cDNA
[0129] Primers were designed to flank the putative open reading frame and used in a
SEQ.ID.NO.:3.:C-E F / L-U 5′-GGATAAAACCTGGGGGGACCTG-3′SEQ.ID.NO.:4:C-E F / L-L 5′-TCCGGGGCCCCAGAGGTAGATGAG-3′
preparative PCR reaction using human prostate marathon ready cDNA (Clontech, Pal...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Composition | aaaaa | aaaaa |
| Resonance energy | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com