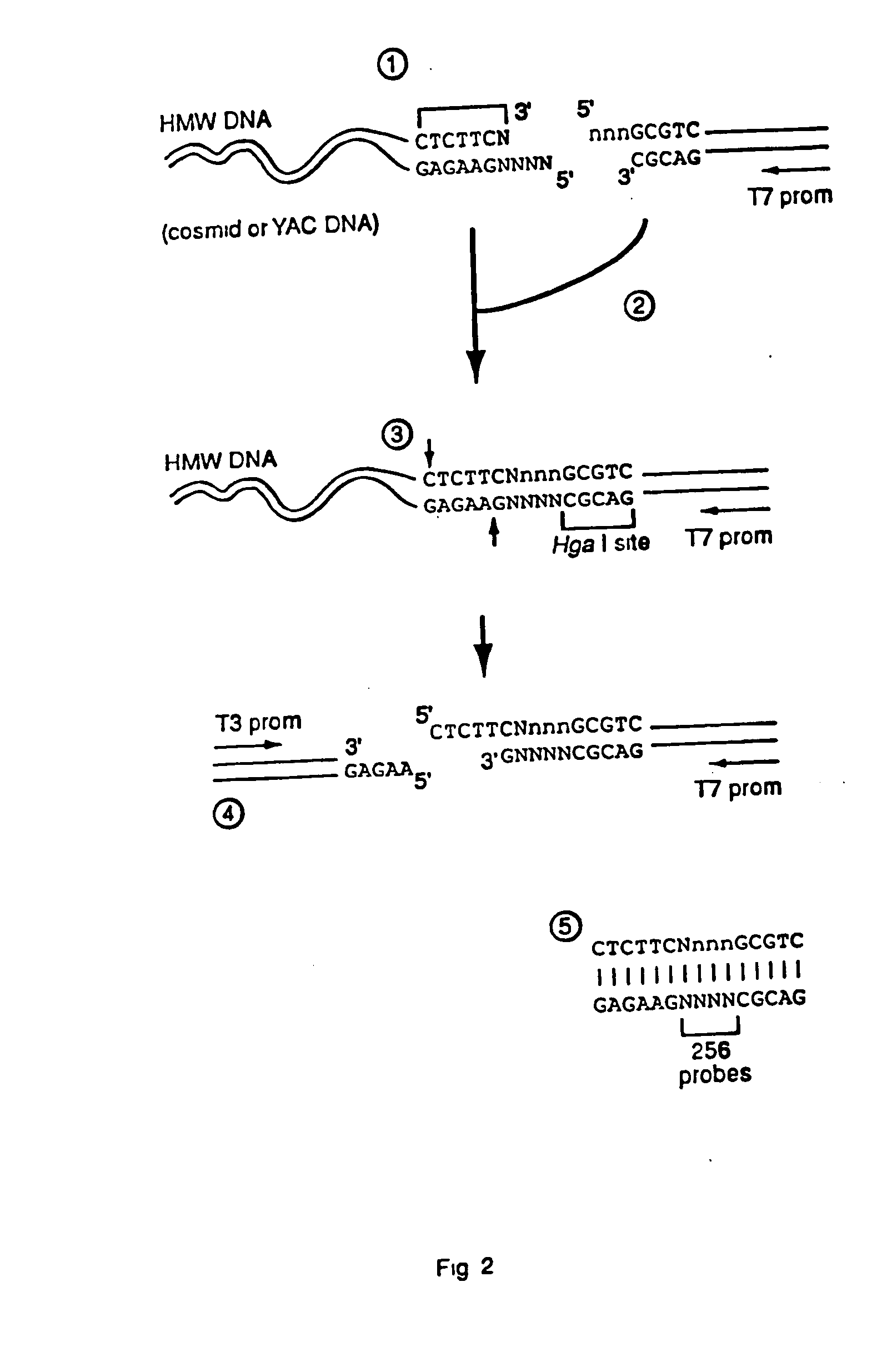
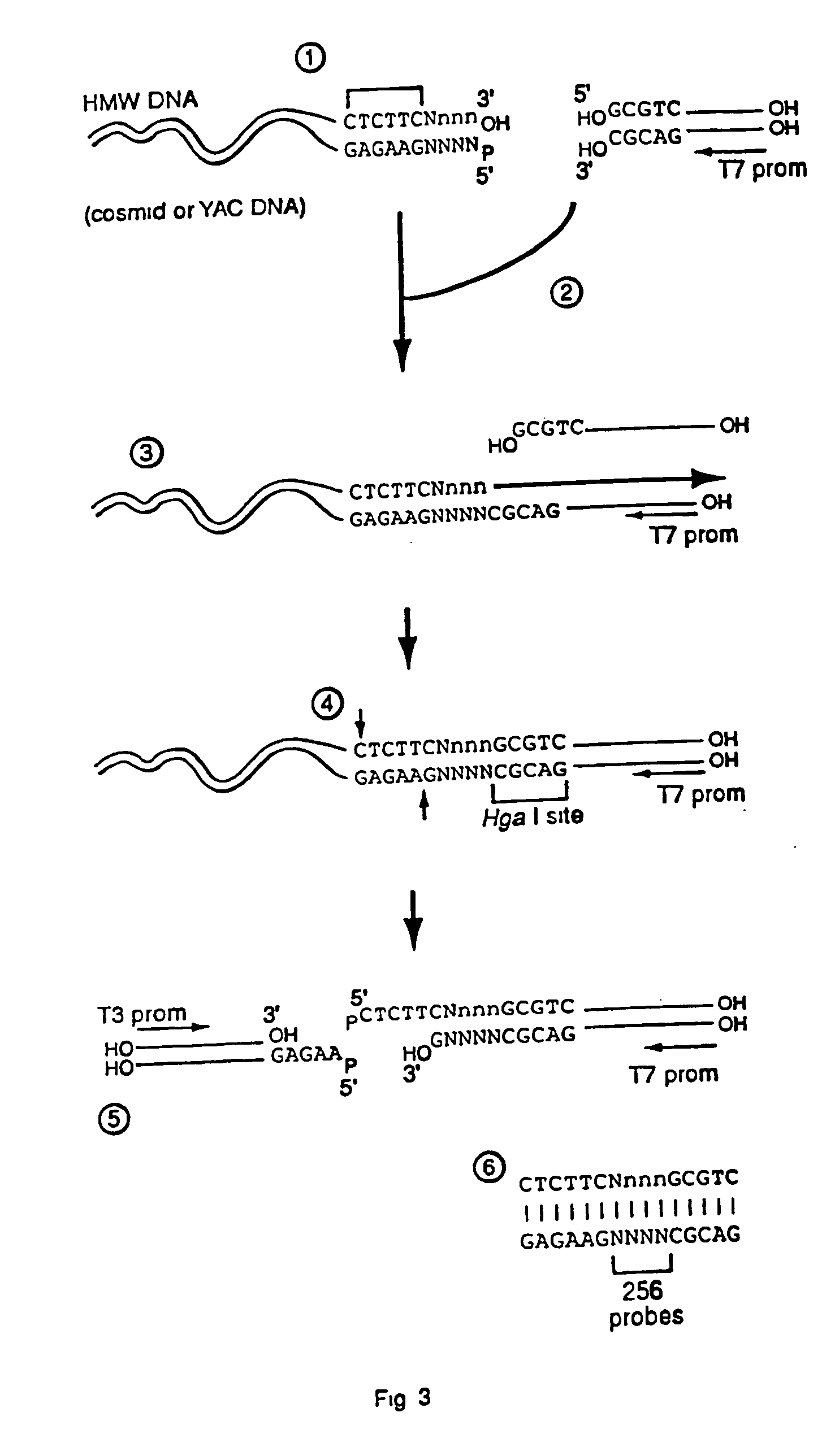
Capturing sequences adjacent to type-IIS restriction sites for genomic library mapping
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
1. Digesting High Molecular Weight DNA with EarI
[0084] 4 μg of λ DNA was treated with 4 units of EarI in 10 μl at 37° C. for 4 hours. The reaction was then heated to 70° C. for 10 minutes. Cleavage was verified by running 5 μl of the sample on an agarose gel to determine complete cleavage. The remaining 5 μl was brought to 40 μl (final concentration of 50 ng / μl λ DNA).
2. Klenow Fill-In Reaction
[0085] 4 μl of the digested λ DNA was added to 0.5 μl of 10× Klenow Buffer, 0.5 μl 2 mM dNTPs, and 0.05 μl of 0.25 units of Klenow fragment. The reaction mixture was incubated for 20 minutes at 25° C., followed by 10 minutes at 75° C. Similar results were also obtained using T4 DNA polymerase for the fill-in reaction.
3. Preparing Adapter Sequences
[0086] Two separate adapter sequences were prepared, adapter sequence 1 and adapter sequence 2. Adapter sequence 1 is used in the first ligation reaction whereas adapter 2 is used for the second. As each adapter and its ligation are somewhat d...
example 2
[0104] The above capture methods were applied to a genomic library of 12 known cosmids from yeast chromosome IV. The clones have been previously physically mapped using EcoRI-HindIII fragmentation. The specific library, including known zap positions and overlap of the 12 cosmids, is illustrated in FIG. 6.
[0105] The twelve genomic clones were constructed in a pHC79 vector, in E. coli host HB101. Cosmid DNA was prepared from 3 ml cultures by an alkaline lysis miniprep method. The miniprep DNA was digested with EcoRI and HindIII to confirm the known fingerprint of the large cloned inserts. Cosmid DNA was treated with linear DNAase, Plasmid-Safe™ DNAse, at 37° C. for 15 minutes, followed by heat inactivation. The DNAse treatment was carried out to remove any potential spurious EarI digested sites resulting from contaminating bacterial DNA. This leaves cosmid DNA substantially untouched. After confirming the presence of clean banding cosmid DNA, the resulting cosmids were then subjected...
example 3
[0109] The correlation scores from yeast chromosome IV, above, were used to construct a best fitting contig, using the simulated annealing process as described by Cuticchia, et al., The use of simulated annealing in chromosome reconstruction experiments based on binary scoring, Genetics (1992) 132:591-601. A global maximum was sought for the sum of correlation coefficient scores for a given sequence of cosmids in the randomly constructed and permutated contig. The resulting high scoring contigs for all 12 cosmids and for the 10 “strong-signal” cosmids are shown below. Each cosmid was assigned a rank based upon the known position of that cosmid, and these are as follows:
TABLE 1Cosmid NumberCosmid Rank9371A8552B808719481298583958348024582536950979460880649983110
[0110] Simulated annealing of all twelve cosmids produced the following ordering:
(1 2 3 4) (7 6 5) A B (8 9 10)
[0111] Inclusion of the weaker signal cosmids, A and B, results in some shuffling of the pr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com