Proteases and methods for producing them
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
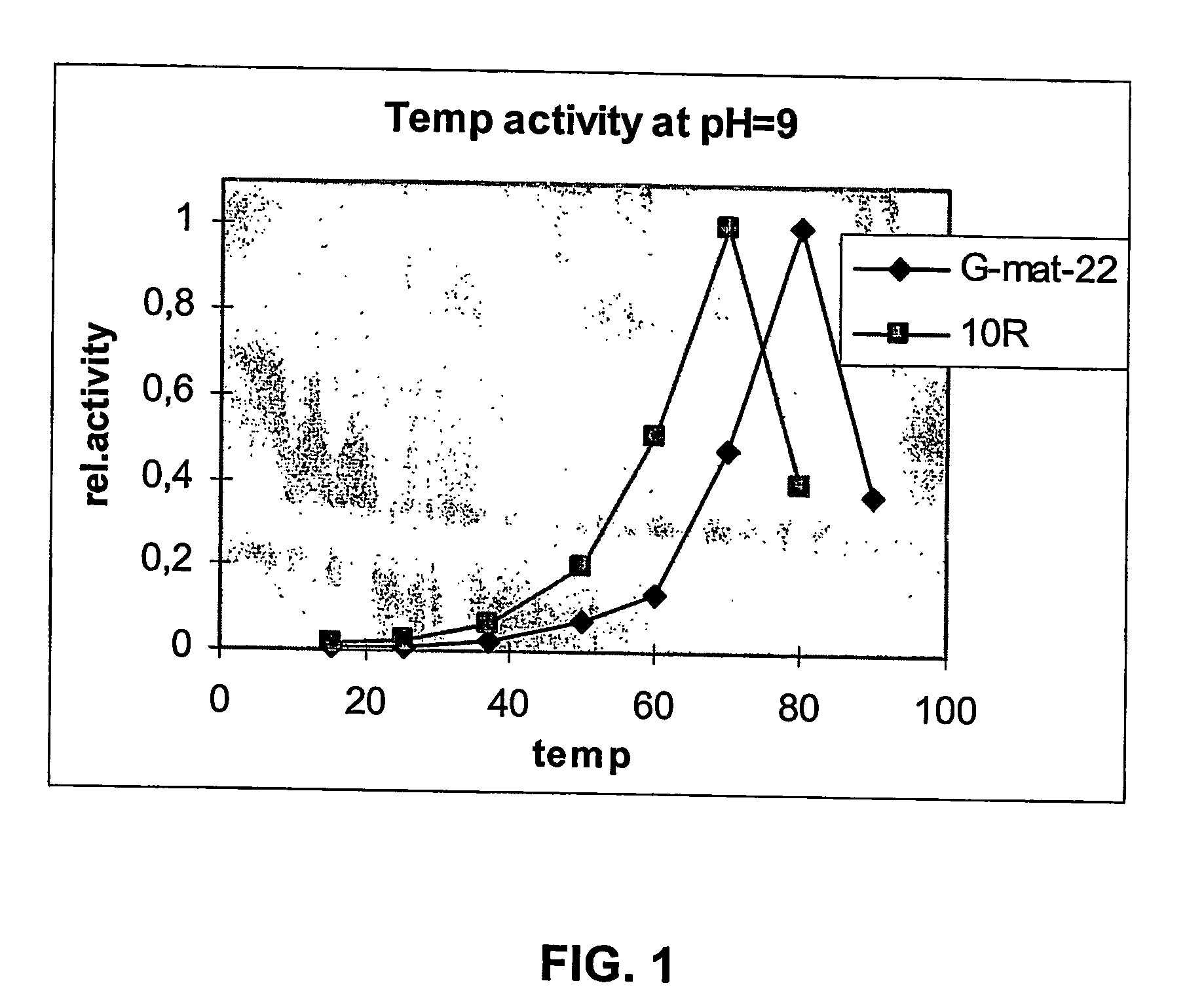
Image
Examples
example 1
Construction of Synthetic 10R Tail-Variant Genes With Savinase Signal
[0280] A synthetic 10R gene (10RS) encoding a S2A protease denoted 10R from Nocardiopsis sp. NRRL 18262 (WO 01 / 58276) was constructed which has the nucleotide sequence shown in SEQ ID NO: 1. This synthetic gene was fused by PCR in frame to the DNA coding for the signal peptide from SAVINASE™ (Novozymes) resulting in the coding sequence Sav-10RS which is shown in SEQ ID NO: 2. Several tail-variants of this construct were made. Compared to the Sav-10RS protease encoded by SEQ ID NO:2 the tail variant construct Sav-10RS HV0 was constructed to have 8 amino acids extra in the C-terminus: QSHVQSAP (SEQ ID NO: 3) which were encoded by the following DNA sequence extension inserted in front of the TAA stopcodon of SEQ ID NO: 2:
[0281] (SEQ ID NO: 4): caatcgcatgttcaatccgctcca
[0282] Tail variant Sav-10RS HV1 was constructed to have 4 amino acids extra in the C-terminus: QSAP (SEQ ID NO: 5), with the following DNA sequence e...
example 2
Fermentation Yields of 10R Tail-Variants With Savinase Signal
[0289] Fermentations for the production of the tail-variant enzymes of the invention were performed on a rotary shaking table in 500 ml baffled Erlenmeyer flasks each containing 100 ml TY supplemented with 6 mg / l chloramphenicol.
[0290] Six Erlenmeyer flasks for each of the five B. subtilis strains from example 1 were fermented in parallel. Two of the six Erlenmeyer flasks were incubated at 37° C. (250 rpm), two at 30° C. (250 rpm), and the last two at 26° C. (250 rpm). A sample was taken from each shake flask at day 1, 2 and 3 and analyzed for proteolytic activity. The results are shown in tables 1-3. As it can be seen from tables 1-3, the effect of the tails is a surprisingly high improvement on the expression level of the protease, as measured by activity in the culture broth. The effect is most pronounced at 26° C. and 30° C., but is also evident at 37° C. as an effect observed especially at the early stage of the fer...
example 3
Chromosomal Integration of Tail-Variant Genes
[0293] The following construct was used for the chromosomal integration of the tail-variant encoding genes. The coding sequence of the well-known subtilisin BPN′ protease was operationally linked to a triple promoter, a marker gene was fused to this (a spectinomycin resistance gene surrounded by resolvase res-sites), and pectate lyase encoding genes from Bacillus subtilis were fused to the construct as flanking segments comprising the 5′ polynucleotide region upstream [yfmD-ytmC-yfmB-yfmA-Pel-start], and the 3′ polynucleotide region downstream [Pel-end-yflS-citS(start)] of the tail-variant encoding polynucleotide, respectively. The integrational cassette was made by the joining of several different PCR fragments. After the final PCR reaction the PCR product was used for transformation of naturally competent B. subtilis cells. One done denoted PL3598-37 was selected and confirmed by sequencing to contain the correct construct.
[0294] The ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com