Genetic markers for skatole metabolism
a skatole and gene technology, applied in the field of gene markers for skatole metabolism, can solve the problems of insufficient explanation of the variation in fat skatole concentration in pigs, and achieve the effects of reducing boar taint, enhancing skatole metabolism, and enhancing aldehyde oxidase activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of Skatole Metabolites
Materials and Methods
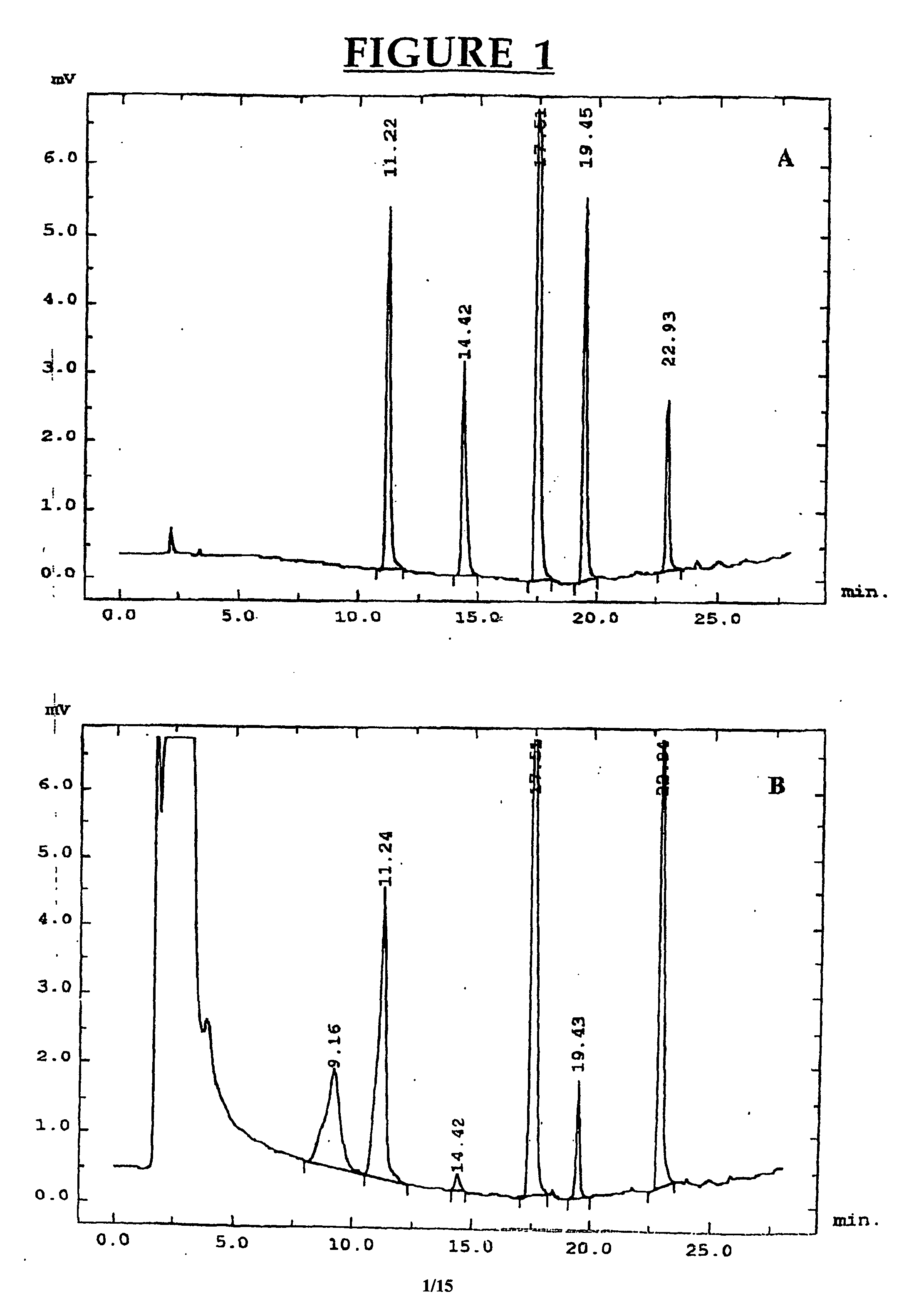
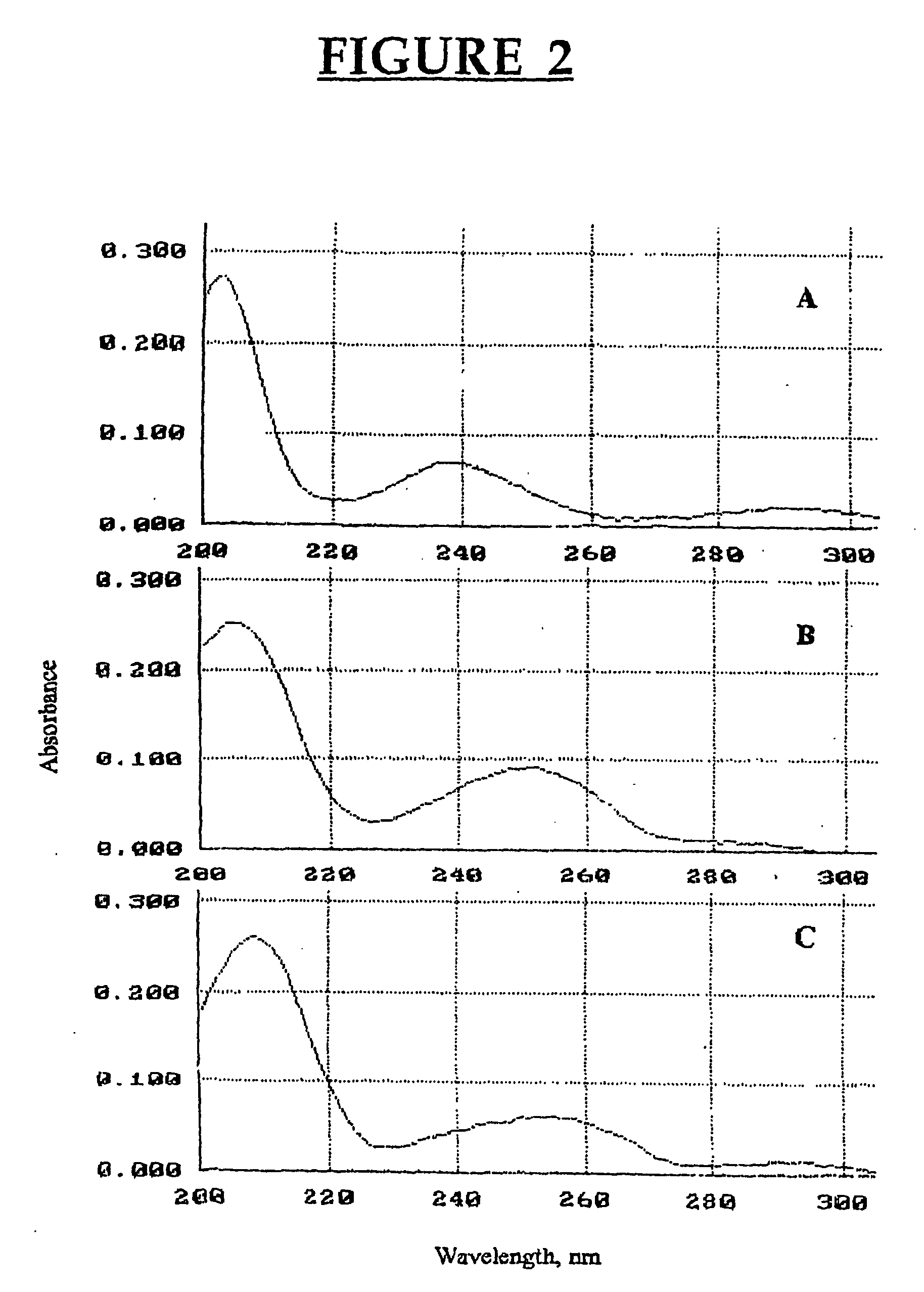
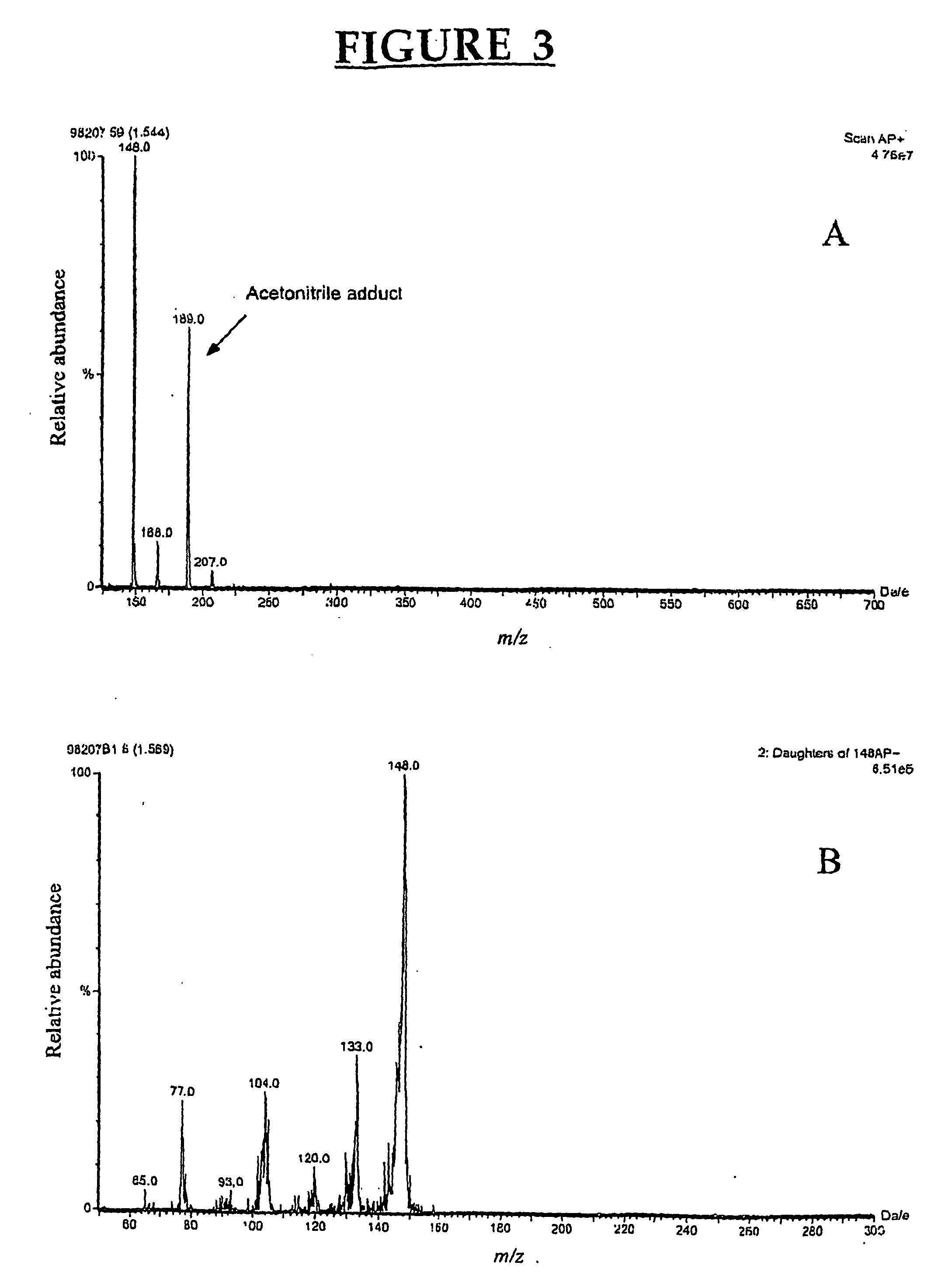
[0085] Chemicals. 3-Methylindole (3MI), indole-3-carbinol (13C), indole-3-aldehyde, indole-3-carboxylic acid, 2-aminoacetophenone and sulfatase type H-2 from Helix pomatia were purchased from Sigma-Aldrich Canada Ltd. (Oakville, ON, Canada). The oxindoles, 3-methyloxindole (3MOI) and 3-hydroxy-3-methyloxindole (HMOI) were synthesized by the methods of Kende and Hodges (1982) and Skiles et al. (1989), respectively. Authentic 5-OH-3-methylindole and 6-OH-3-methylindole (in the form of 6-sulfatoxyskatole) were donated by Jens Hansen-Møller (Danish Meat Research Institute, Roskilde, Denmark). In order to obtain 6-OH-3-methylindole from 6-sulfatoxyskatole, the compound was hydrolyzed in a total volume of 0.5 ml acetate buffer pH 5.0 containing 90 units / ml of type H-2 sulfatase. Hydrolysis was conducted for 4 hours in a shaking water bath at 40° C. and then 0.5 ml of ice-cold acetonitrile were added both to stop the reaction an...
example 2
Aldehyde Oxidase
Materials And Methods
[0104] Chemicals. Menadione, quinacrine and allopurinol were purchased from Sigma-Aldrich Canada (Oakville, ON, Canada). Authentic HMOI was graciously provided by Dr. G. S. Yost, Department of Pharmacology and Toxicology, University of Utah. HMI was produced using porcine liver microsomes and it was isolated and purified using preparative HPLC as described before (Diaz et al., 1999). Isolated HMI was freeze-dried and kept in a dessicator at −20° C. until used.
[0105] Preparation of porcine liver cytosol. Liver samples were taken from 30 intact male pigs obtained by back-crossing F3 European Wild Pig×Swedish Yorkshire boars with Swedish Yorkshire sows (Squires and Lundström, 1997). Liver samples were frozen in liquid nitrogen and stored at −80° C. For the preparation of the cytosolic fraction, partially thawed liver samples were finely minced and homogenized with 4 volumes of 0.05 M Tris-HCl buffer pH 7.4 (containing 0.15 M KCl, 1 mM EDTA, and...
example 3
The Role of CYP2A6 IN 3-Methylindole Metabolism by Porcine Liver Microsomes
[0118] The role of different cytochrome P450 enzymes on the metabolism of 3-methylindole (3MI) was investigated using selective chemical inhibitors. Eight chemical inhibitors of P450 enzymes were screened for their inhibitory specificity towards 3MI metabolism in porcine microsomes: alpha-naphthoflavone (CYP1A2), 8-methoxypsoralen (CYP2A6), menthofuran (CYP2A6), sulphaphenazole (CYP2C9), quinidine (CYP2D6), 4-methylpyrazole (CYP2E1), diethyldithiocarbamate (CYP2E1, CYP2A6), and troleandomycin (CYP3A4). The production of the different 3MI metabolites was only affected by the presence of inhibitors of CYP2E1 and CYP2A6 in the microsomal incubations. In a second experiment, a set of porcine microsomes (n=30) was screened for CYP2A6 content by Western blot analysis and also for their 7-hydroxylation activity (CYP2A6 activity). Protein content and enzymatic activity were found to be correlated with 3MI fat conte...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| total volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com