Purified active HCV NS2/3 protease
a technology of hepatitis c virus and ns2/3, which is applied in the direction of peptides, chemical treatment enzyme inactivation, enzymology, etc., to achieve the effect of promoting autocleavage and reducing difficulties and disadvantages
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
NS2 / 3 Full Length Construct:
[0156] The full length (810-1206) NS2 / 3 sequence was amplified by PCR from the HCV 1b-40 sequence (WO 99 / 07733 by Boehringer-Ingelheim (Canada), Ltd.) using two oligonucleotide primers, 5′-CCATGGACCGGGAGATGGCT-3′ (SEQ ID NO: 5) for the N-terminus and 5′GGATCCTTAACCACCGAACTGCGGGTGACGCCAAGCGCTACTAGTCCGCAT GGTAGTTTCCAT-3′ (SEQ ID NO: 6) for the C-terminus. This procedure introduces a NcoI site at the 5′ end and a streptavidin tag “st” (Schmidt & Skerra, Prot. Engineering (1993) 6; 109-122) followed by a BamH1 site at the 3′ end giving a nucleic acid molecule of SEQ ID NO:1 (FIG. 1). The PCR product was inserted into the vector pCR™ 3 using the TA cloning® kit from Invitrogen. The insert was then transferred to a bacterial expression vector pET-11d (Novagen) by cutting with EcoRI followed by Klenow treatment to create blunt ends followed by a partial digestion with NcoI. This construct was designated pET-11d-NS2 / 3st. The DNA was transformed into XL-1 Blue E...
example 2
NS2 / 3 N-terminal Deletion Mutants
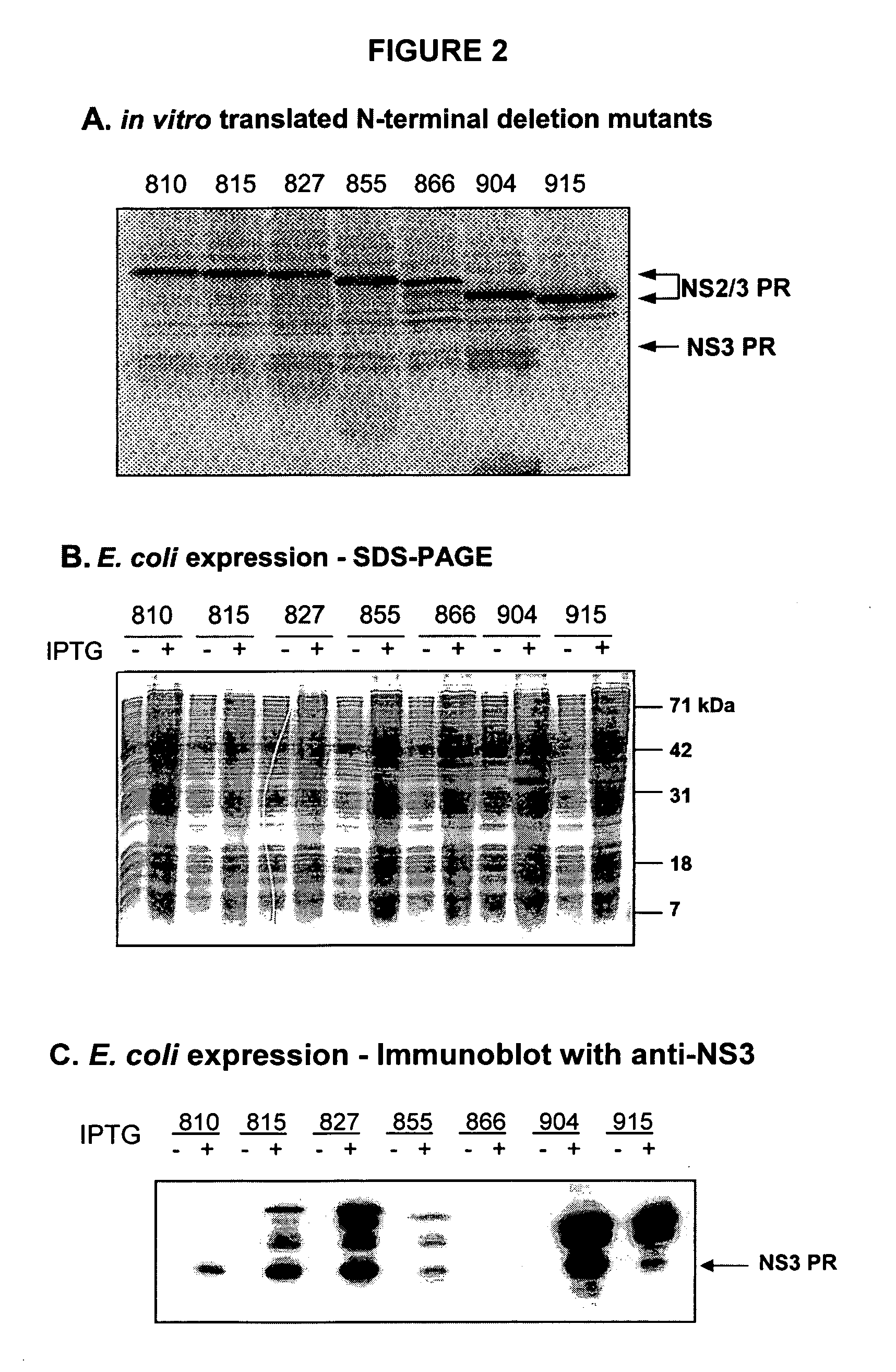
[0157] The N-terminal deletion mutants 815*-1206, 827-1206, 855-1206, 866-1206, 904-1206 and 915-1206 were derived from the pET-11d / NS2 / 3 template that was designed with a NcoI site at the 5′ end and within the NS3 domain at amino acid 1083. Following the template digestion with NcoI, the 3′ end fragment and the vector were gel purified. The mutants were obtained by PCR using the appropriate synthetic oligonucleotides primers containing the NS2 / 3 sequence from nucleotides that encode the desired N-terminal residue up to amino acid 1083. The primers also introduced a NcoI site at the 5′ end, such that the resulting inserts could be ligated to the gel purified fragment. The DNA was then transformed into E. coli XL-Blue cells, isolated and sequenced. Finally, the DNA was transferred into E. coli BL21(DE3)pLysS for protein production. Expression was verified by SDS-PAGE (FIG. 2A, 2B, 2C).
* The numbering of this fragment is erroneous since the first me...
example 3
NS2 / 3 N-terminal Truncation Mutant 4K-6H (904-1206)st-4K (SEQ ID NO: 4):
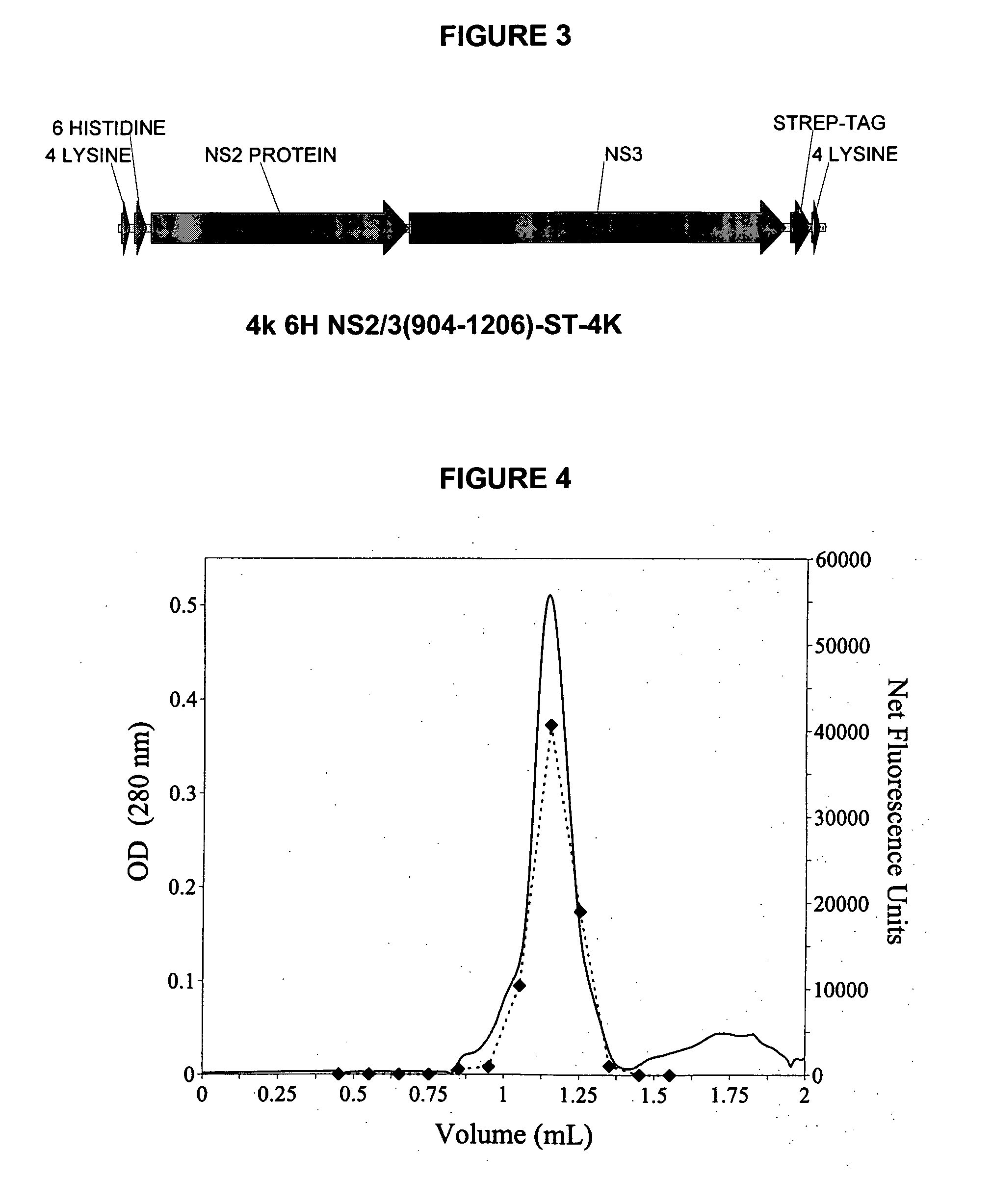
[0158] In this construct, four lysines were added at the N- and C-termini as well as a hexahistidine tag at the N-terminus. This construct was obtained using PCR and the pET-11d / NS2 / 3st template with two primers containing the sequence for the tags as well as the NS2 / 3 sequence from nucleotides that encode amino acid residues 904-1206. The primers also introduced a NdeI and BamHI site at 5′ and 3′ end respectively. The insert was cloned into pET-11d and designated pET-11d 4K-6H-NS2 / 3 (904-1206)-st-4K (SEQ ID NO:3). The DNA was transformed into E. coli XL-1 Blue cells, isolated and sequenced. The DNA was then transferred into E. coli BL21 (DE3) pLysS for protein production.
[0159] For the truncated construct 904-1206, 4 primers and 2 successive PCR reactions were used. The primers used in the first PCR reaction were GCTCGAGCATCACCATCACCATCACACTAGTGCAGGCATAACCAAA (SEQ ID NO:7) for the N-terminus and AACAATGGATCC...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com