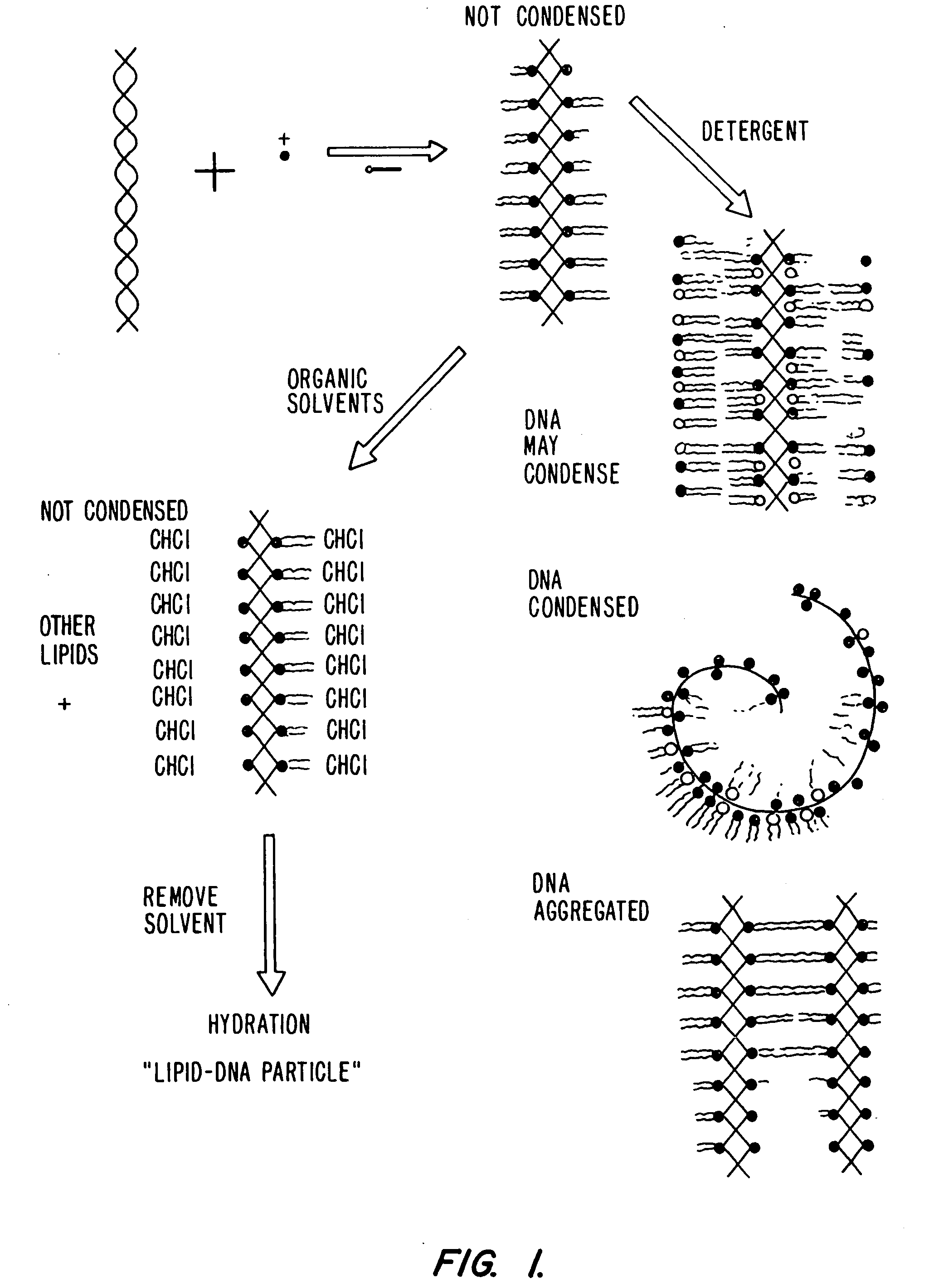
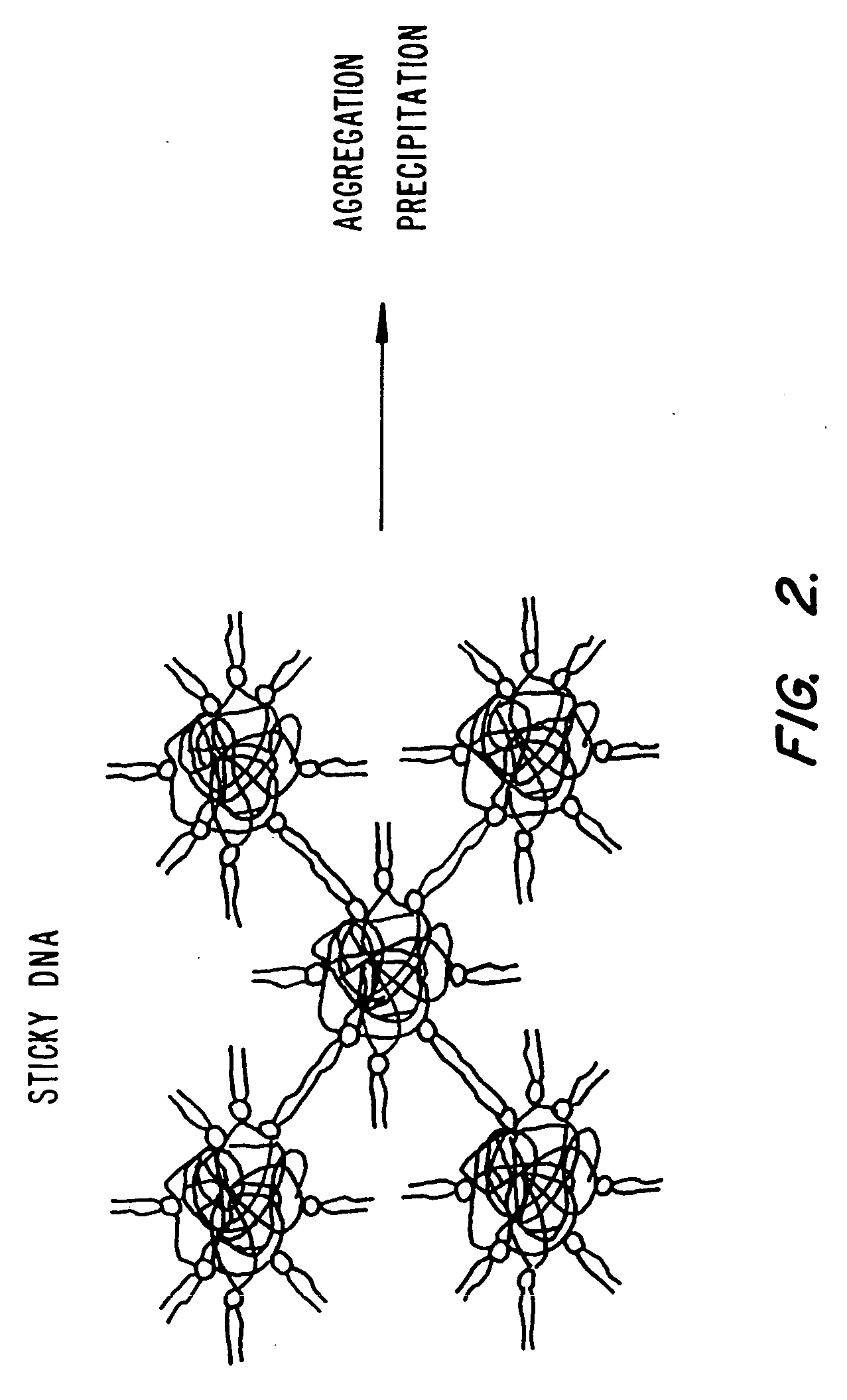
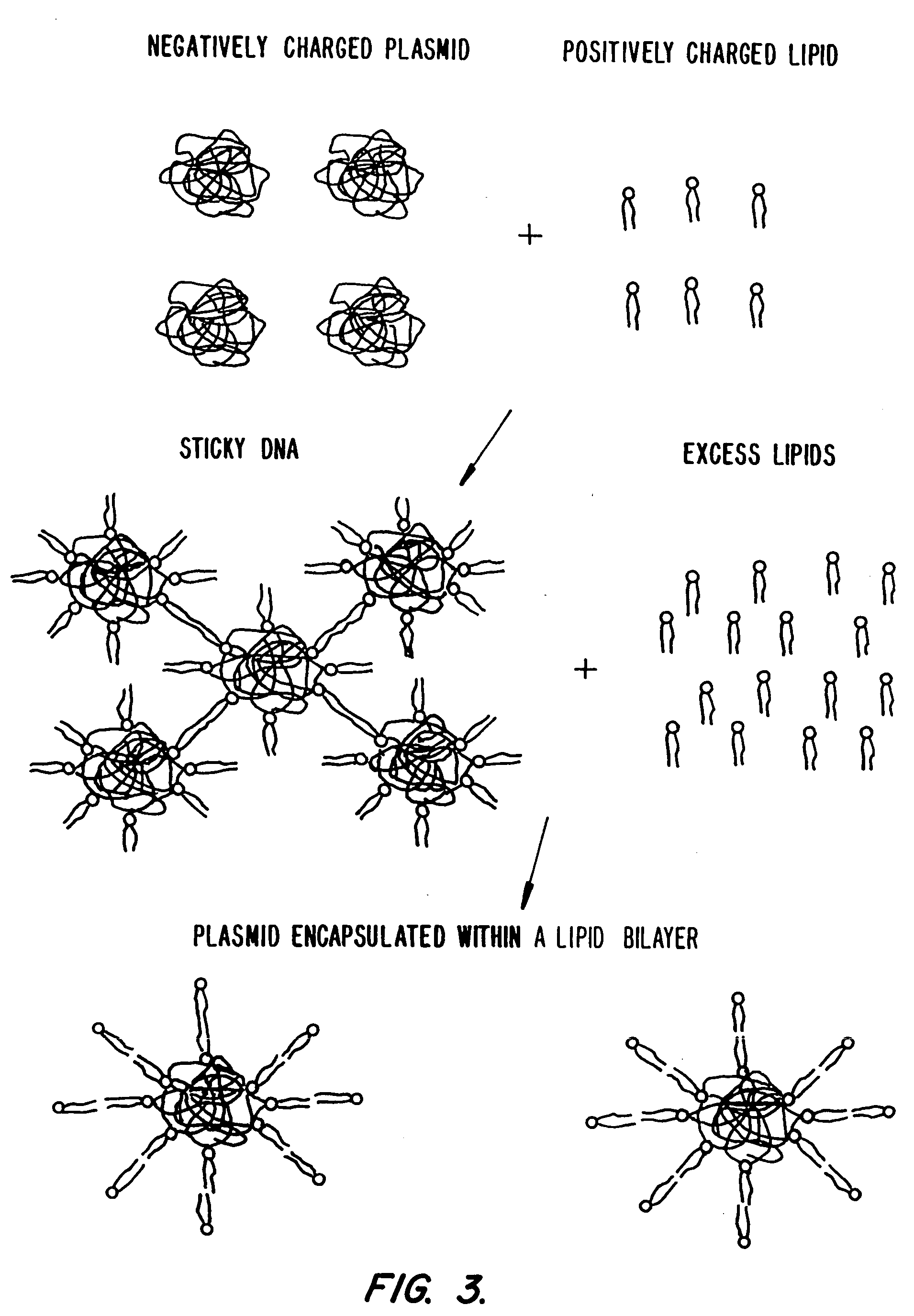
Lipid-nucleic acid particles prepared via a hydrophobic lipid-nucleic acid complex intermediate and use of gene transfer
a technology of lipid-nucleic acid complex and intermediate, which is applied in the field of lipid-nucleic acid particles, can solve the problems of complex gene transfer, limited dna carrying capacity, and inability to sel
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0243] This example illustrates the encapsulation of a plasmid in a lipid particle using either a reverse-phase method or a detergent dialysis method.
Reverse Phase Method
[0244] pCMV4-CAT plasmid was encapsulated in a lipid particle which was constructed using about 10 mg or 20 mg of lipid. The encapsulation method involved a modification of the classical reverse phase method for entrapment. Generally, 1.050 ml of chloroform: methanol in a 1:2-1 mole % ratio was added to a lipid film containing 2 μl of 14C-cholesteryl hexadecyl ether (6.66 μl / μCi). This was followed by the addition of 220 μl H2O and 33 μl 3H-pCMVCAT plasmid (158,000 dpm / μl; 1.5 mg / ml). This combination provided a clear single phase. The chloroform and most of the methanol were removed under a stream of nitrogen while vortexing. In some cases, the resulting 250 μl suspension of encapsulated plasmid was diluted with 1 ml of H2O and extruded 5 times through one 400 nm filter followed by extrusion 5 times through one ...
example 2
[0246] This example illustrates the level of plasmid “protection” from the external medium using anion exchange chromatography.
[0247] The extent of encapsulation or protection of the plasmid from the external medium was assessed by anion exchange chromatography as follows: a 50 μl aliquot of each sample was eluted on a DEAE Sepharose CL-6B column and the fractions were assessed for both 3H-plasmid and 14C-lipid by scintillation counting. Any exposed negative charges, such as those present on DNA molecules will bind to the anion exchange column and will not elute with the 14C-lipid. DNA which has its negative charge, “protected” or non-exposed will not bind to the anion exchange resin and will elute with the 14C-lipid. Alternatively, plasmid DNA was measured using the indicator dye, PicoGreen®.
Reverse Phase Method
[0248]FIG. 4 presents the results describing the relationship between the amount of DODAC present in the formulation and the encapsulation efficiency for POPC:DODAC:PEG-...
example 3
[0250] This example illustrates the serum stability achieved using plasmid:lipid particles prepared by the methods of Example 1.
[0251] To establish the serum stability of the plasmid-lipid particles aliquots of the particle mixtures prepared according to both the reverse phase and dialysis method of Example 1 were incubated in 80% mouse serum (Cedar Lane) for 30 min at 37° C. Prior to incubation, the lipid associated plasmid was eluted on a DEAE Sepharose CL-6B column to remove unencapsulated plasmid. Following incubation, an aliquot of the incubation mixture was eluted in HBS on a Sepharose CL-4B column.
[0252] As a control, 1.5 mg of free 3H-pCMVCAT was eluted on a Sepharose CL-4B column in HBS, pH 7.4 (see FIG. 9A). For comparison, 1.5 mg of free 3H-pCMVCAT was incubated in 500 μl of mouse serum at 37° C. for 30 min and eluted in the same manner (FIG. 9B). Note that in FIG. 9A, the free plasmid eluted in the void volume of the column while, in FIG. 9B, the plasmid incubated in s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com