Systems and Methods for Controlling the Position of a Charged Polymer Inside a Nanopore
a charged polymer and nanopore technology, applied in the field of polymer characterization, can solve the problems of slow reagent cycle time (tens of seconds), expensive reagents, and short read length (tens to hundreds of bases)
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
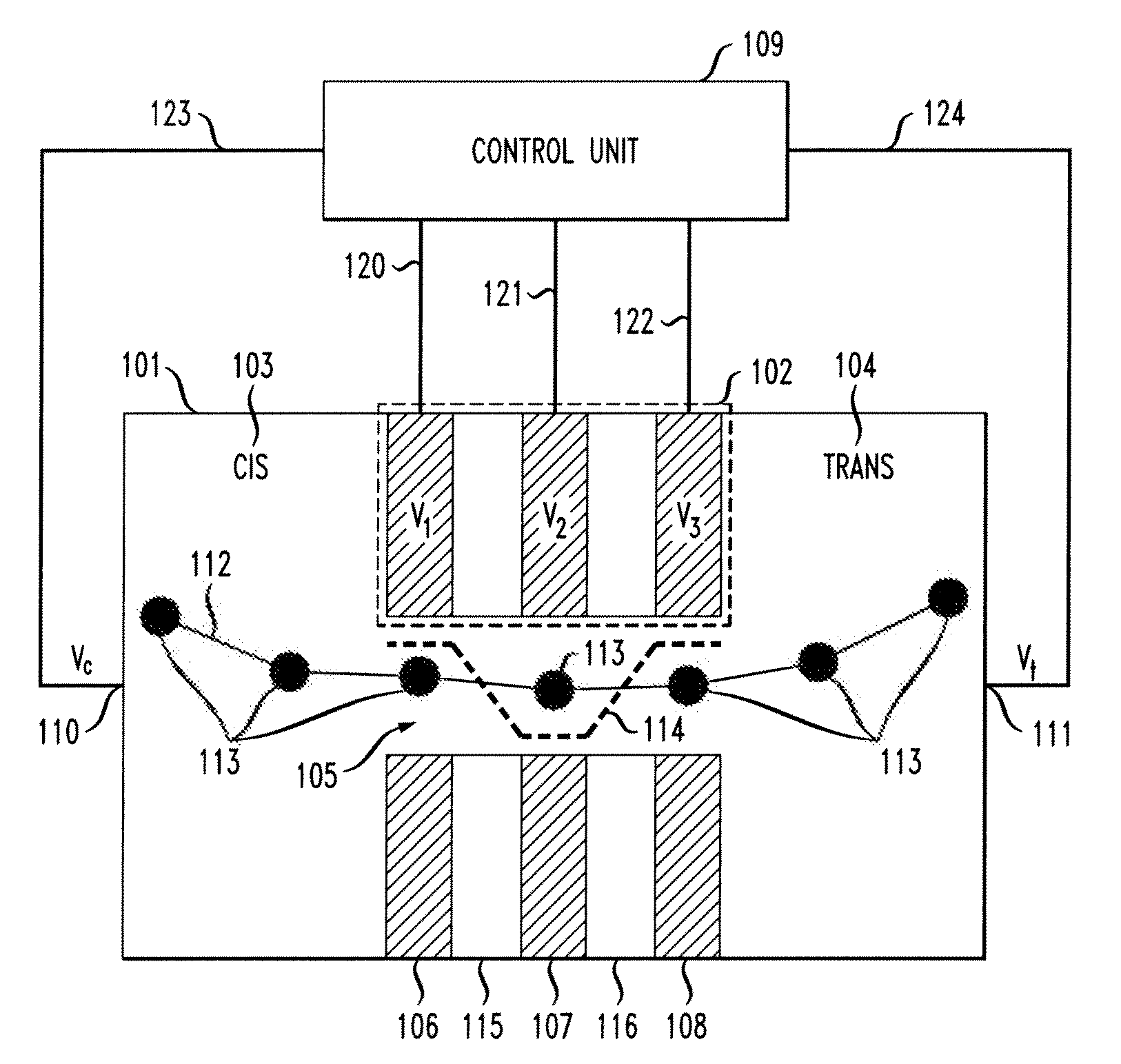
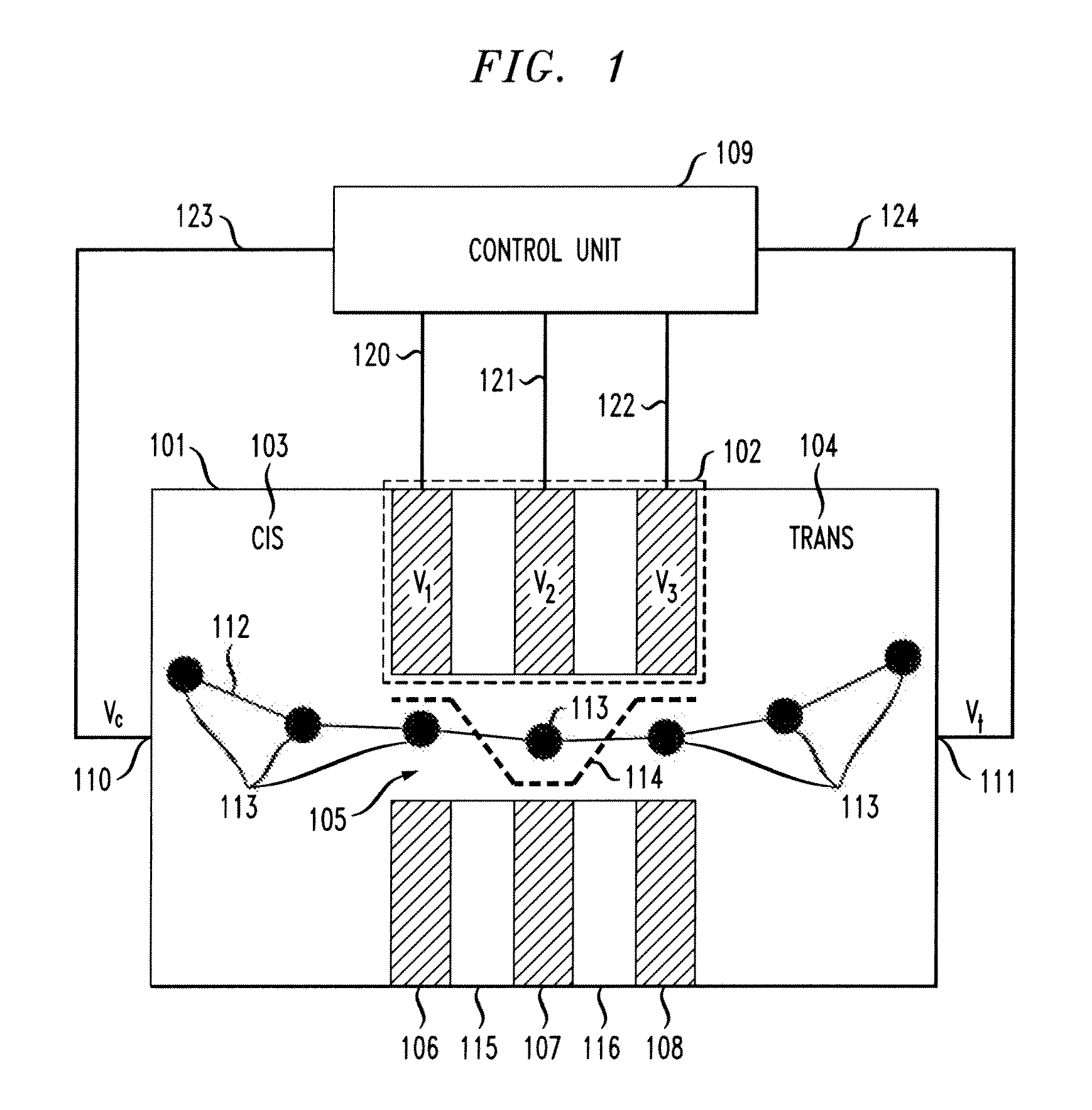
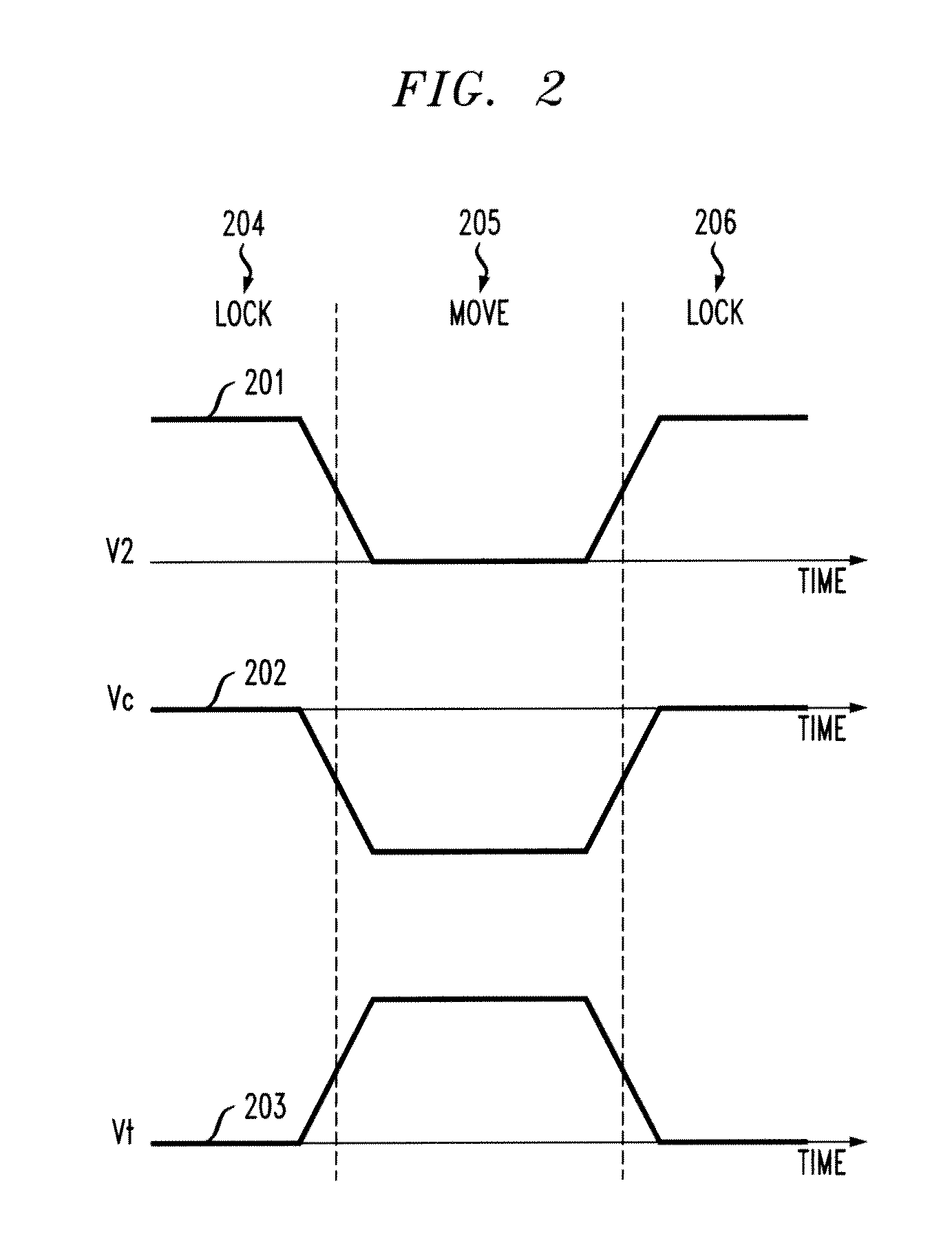
[0022]As noted above, it would be beneficial to not only slow down the translocation of charged polymers, but to control the position of a polymer inside a nanopore with single nucleotide accuracy. Principles of the present invention use an electrostatic potential well to lock the positions of linear polymers carrying localized charges along their chain. Electrostatic control (ESC) is used to position and move a polymer such as, for example, deoxyribonucleic acid (DNA), inside a nanopore.
[0023]Principles of the present invention apply varying voltages to metal layers in order to produce sensitive control of the position of negatively-charged nucleotides. The control may be similar to the charge control in charge-coupled device (CDD) sensors.
[0024]One or more embodiments of the present invention detect nucleotide type by measuring tunnel current of capacitance change between layers. Also, layer voltages may be modulated with high frequency signals in order to utilize lock-in measurem...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameters | aaaaa | aaaaa |
| voltage | aaaaa | aaaaa |
| voltage | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com