Production of commercial biodiesel from genetically modified microorganisms
a technology of genetically modified microorganisms and biodiesel, which is applied in the direction of biofuels, fermentation, biofeedstock, etc., to achieve the effects of low impurities and/or undesirable contaminants, and clean emissions profiles
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0314]This example describes the construction of a genetically engineered microorganism wherein the expression of a fatty acid degradation enzyme is attenuated.
[0315]The fadE gene of E. coli MG1655 (an E. coli K strain) was deleted using the Lambda Red system described in Datsenko et al., Proc. Natl. Acad. Sci. USA 97: 6640-6645 (2000), with the following modifications.
[0316]Two primers were used to create the deletion:
Del-fadE-F(SEQ ID NO: 1)5′-AAAAACAGCAACAATGTGAGCTTTGTTGTAATTATATTGTAAACATATTGATTCCGGGGATCCGTCGACCDel-fadE-R(SEQ ID NO: 2)5′-AAACGGAGCCTTTCGGCTCCGTTATTCATTTACGCGGCTTCAACTTTCCTGTAGGCTGGAGCTGCTTC
[0317]The Del-fadE-F and Del-fadE-R primers were used to amplify the Kanamycin resistance (KmR) cassette from plasmid pKD13 (as described in Datsenko et al., supra) by PCR. The PCR product was then used to transform electrocompetent E. coli MG1655 cells containing pKD46 (described in Datsenko et al., supra). These cells had been previously induced with arabinose for 3-4 h. Follow...
example 2
[0319]This example describes the construction of a genetically engineered microorganism in which the expression of a fatty acid degradation enzyme and an outer membrane protein receptor are attenuated.
[0320]The fhuA (also known as tonA) gene of E. coli MG1655, which encodes a ferrichrome outer membrane transporter (GenBank Accession No. NP—414692), was deleted from strain E. coli MG1655 D1 of Example 1 using the Lambda Red system described in Datsenko et al., supra, but with the following modifications.
[0321]Two primers were used to create the deletion:
Del-fhuA-F(SEQ ID NO: 5)5′-ATCATTCTCGTTTACGTTATCATTCACTTTACATCAGAGATATACCAATGATTCCGGGGATCCGTCGACC;Del-fhuA-R(SEQ ID NO: 6)5′-GCACGGAAATCCGTGCCCCAAAAGAGAAATTAGAAACGGAAGGTTGCGGTTGTAGGCTGGAGCTGCTTC
[0322]The Del-fhuA-F and Del-fhuA-R primers were used to amplify the KmR cassette from plasmid pKD13 by PCR. The PCR product obtained was used to transform the electrocompetent E. coli MG1655 D1 cells containing pKD46 (see Example 1). These cel...
example 3
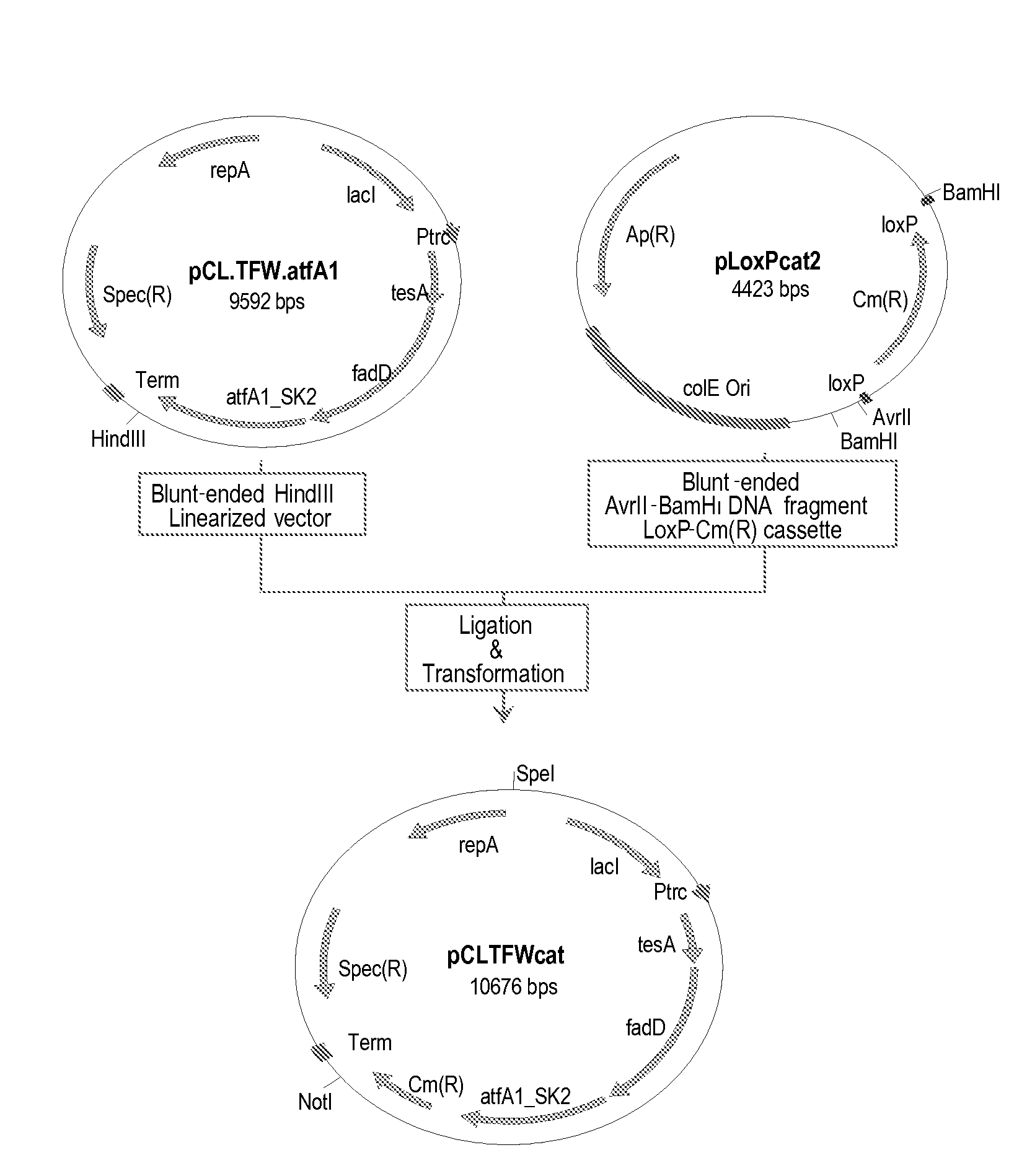
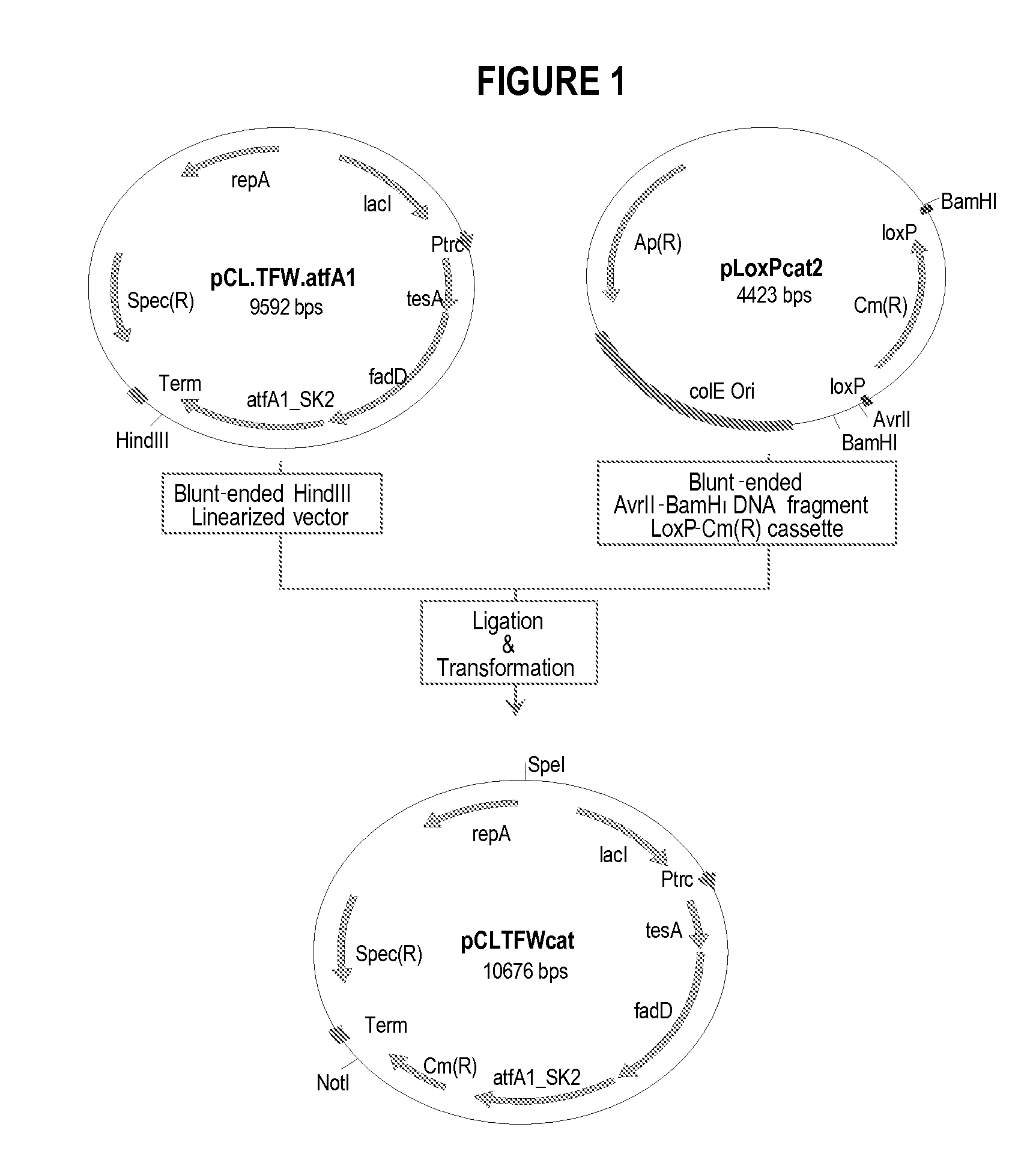
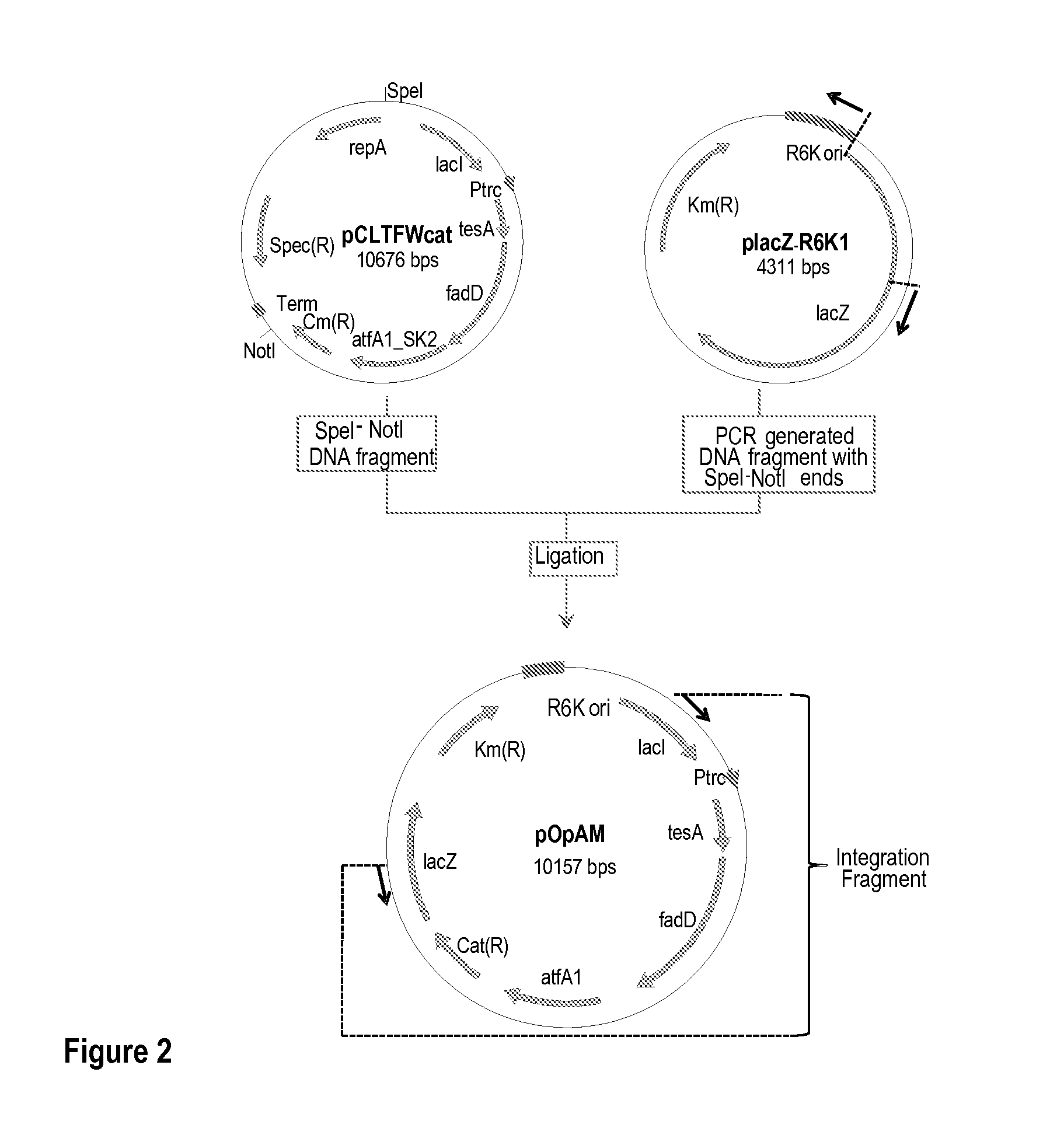
[0325]This example describes the construction of a genetically engineered microorganism in which nucleotide sequences encoding a thioesterase, an acyl-CoA synthase, and an ester synthase are integrated into the microorganism's chromosome.
[0326]The following nucleotide sequences, ′tesA, fadD, and aftA1, were integrated into the chromosome of E. coli MG1655 ΔfadE ΔfhuA strain (or DV2 strain, see Example 2) at the lacZ locus. The sequences were integrated in the order of ′tesA, followed by fadD, and followed by aftA1.
[0327]′tesA is a nucleotide sequence comprising a leaderless E. coli tesA (GenBank entry AAC73596, refseq accession U00096.2). ′tesA encodes an E. coli thioesterase (EC 3.1.1.5, 3.1.2.-) in which the first twenty-five amino acids were deleted and the amino acid in position 26, alanine, was replaced with methionine. That methionine then became the first amino acid of ′tesA. See Cho et al., J. Biol. Chem., 270:4216-4219 (1995).
[0328]E. coli fadD (GenBank entry AAC74875; REFS...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com