Microorganism for expressing a human membrane protein
a technology of membrane protein and microorganism, which is applied in the field of isolated genetically modified living non-mammal organisms, can solve the problems of heterologous expression of membrane protein from mammal cells in wild type strains, difficulty in expressing sert,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Used Materials and Methods
1.1: Primers.
[0058]All primers were synthesized by the company Metabion (Martinsried).
TABLE 1PCR primers for amplification. The recognitionsequences for the restriction enzymes NotIand XhoI are underlined.PrimerSequencemSERTNotIf5′-ATGCGGCCGCACCATGGAGACCACACCTTTG-3′mSERTXhoIr5′-ATCTCGAGTTACACAGCATTCATGCGG-3′
1.2: Plasmids.
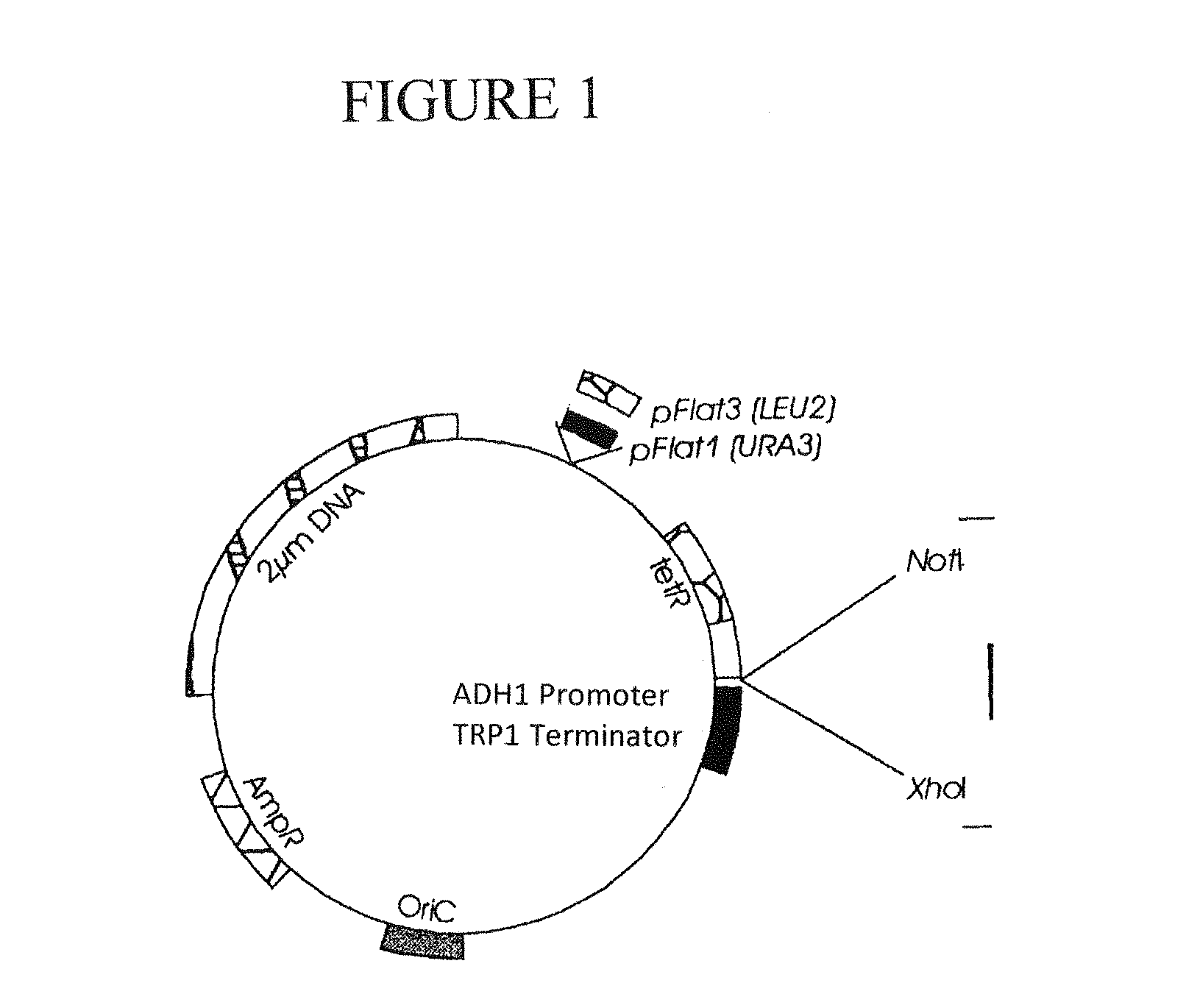
[0059]pFlat vector system (Veen, M., dissertation, TU Berlin (2002)).
[0060]In the following experiments, different strains of the yeast S. cerevisiae were transformed with the pFlat1 or pFlat3 vector of the pFlat vector system (Jacobs B L, Azmitia E C. Physiol Rev (1992) 72, 165-229). These E. coliS. cerevisiae “shuttle vectors” comprise a NotI and a XhoI restriction site in the “Multiple cloning site”, which were used in the present work for cloning. Furthermore, these plasmids carry the LEU2 (pFlat3) or the URA3 gene (pFlat1), respectively, which are used as auxotrophy marker genes for the selection of plasmid-carrying strains on correspo...
example 2
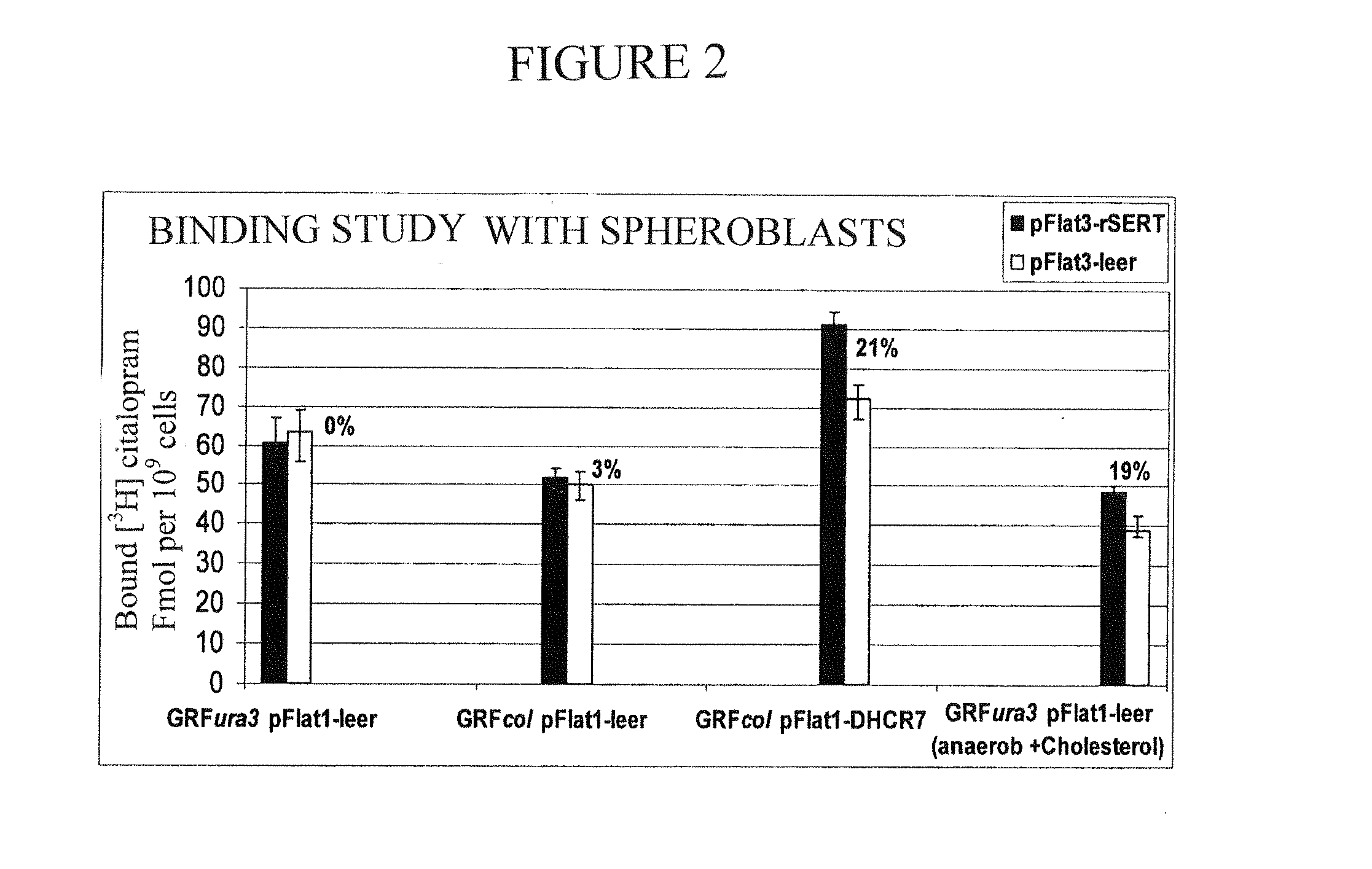
Results
[0092]In the year 2002, the biosynthesis of cholesta-5,7,24-trien-3-ol in the yeast S. cerevisiae could be established (Veen, 2002, see above). For this purpose, a freely obtainable lab strain GRF18 as shown in the following was modified with genetic methods.
StrainDescriptionGRFl8↓leu2, his3GRFura3↓leu2, his3, ura3GRFtH3↓tHMG1-leu2, his3, ura3GRFtH3E1e5↓tHMG1-leu2, ERG1-erg5, his3, ura3GRFtH1E1E11e5e6tHMG1-leu2, ERG1-erg5, ERG11-erg6,his3, ura3
[0093]As can be seen, first the gene URA3 was deleted and the gene tHMG1 was integrated at the gene locus LEU2. The gene tHMG1 (t=truncated, cut short) codes for a cut short HMG-CoA reductase, which only consists of the catalytic subunit of the protein and is thus not subject anymore to the feedback inhibition by sterol intermediates. The transcription of the gene tHMG1 is controlled by the constitutive ADH1 promoter. By the analysis of a systematic overexpression and transcriptional deregulation of all genes of the post-squalene biosyn...
PUM
| Property | Measurement | Unit |
|---|---|---|
| voltage | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com