System and Methods for Massively Parallel Analysis of Nucleic Acids in Single Cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Methods of T-cell Analysis
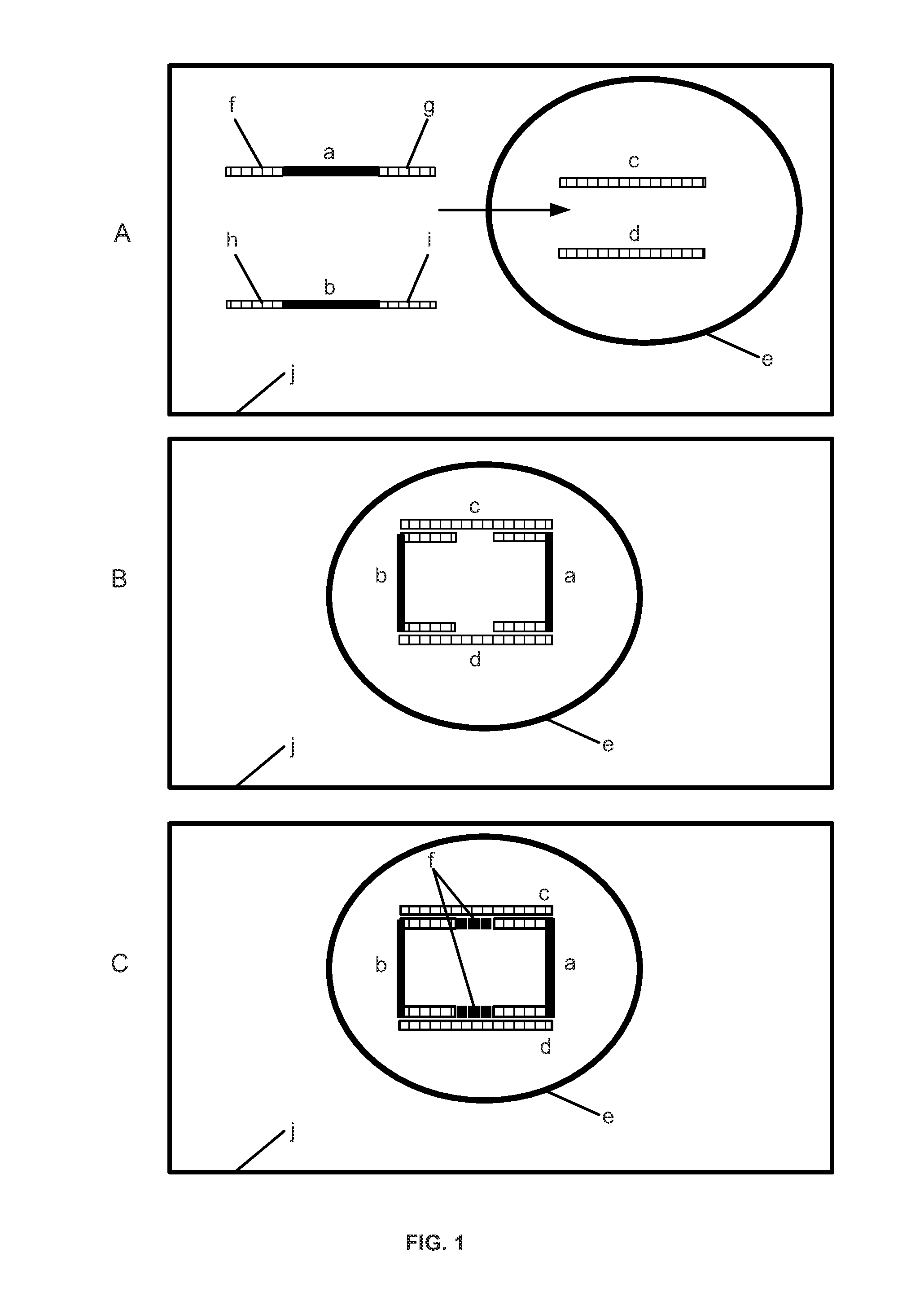
[0184]The immune system responds to disease by inducing cellular responses. Nearly all immunology is involved with detection of clonotype expansion or contraction in response to an antigen and / or functional analysis of the expanded or contracted clonotypes. Described in this example are methods that leverage the information contained in immune response to diagnose and treat disease. Active and / or memory cells are particularly informative because these cells indicate a functional immune response to a disease, and therefore have high information content. Variable DNA regions and RNA transcripts were analyzed in single cells from populations of activated and / or memory immune cells, and then correlated with disease. These profiles were used to develop noninvasive diagnostics, high-value diagnostics that inform treatment regimens, and novel therapeutic agents.
[0185]T cells include T cell receptors (TCR) that recognize antigens and control immune responses. The T...
example 2
High-Throughput Protocol for TCRβ Repertoire Library Construction
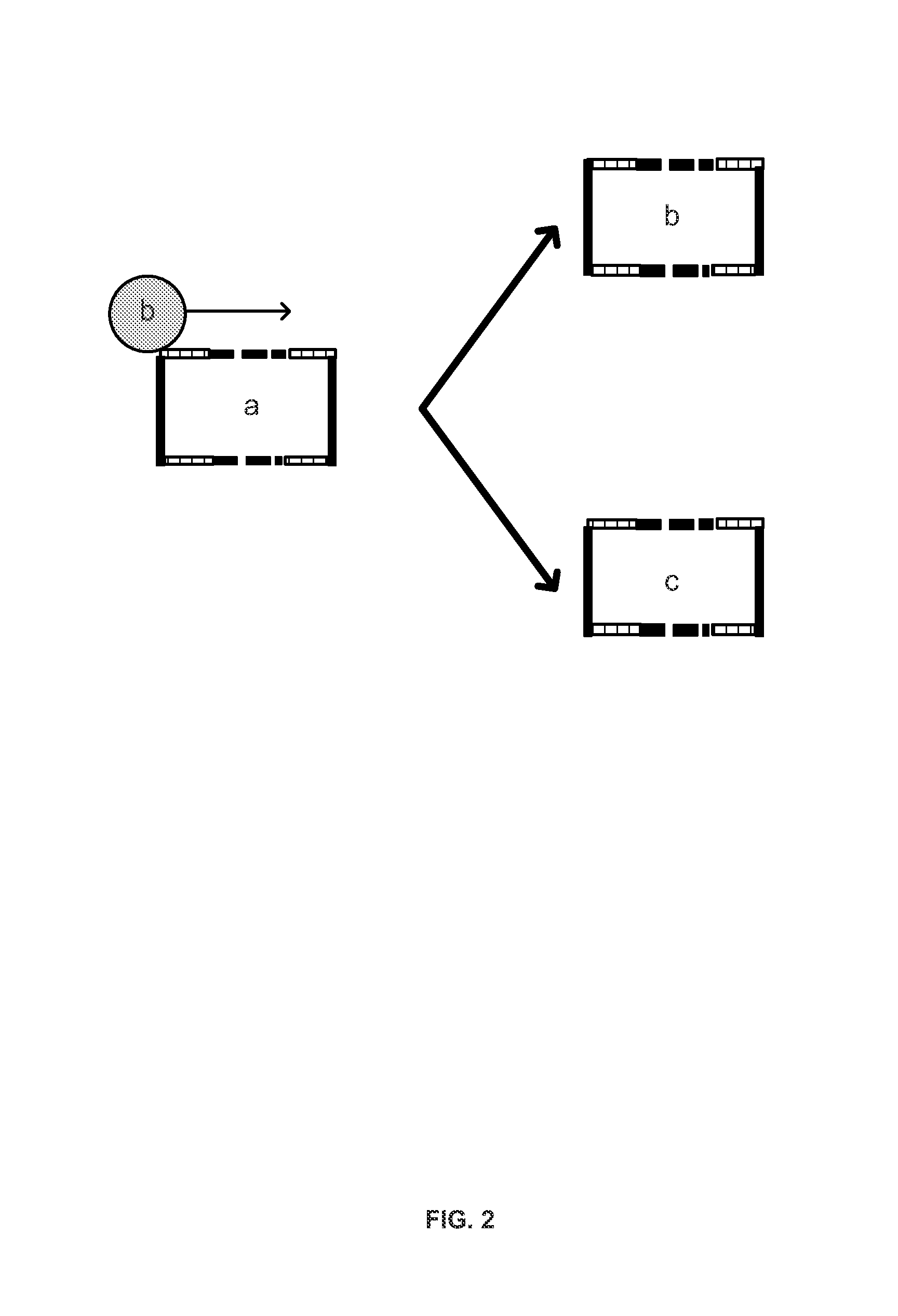
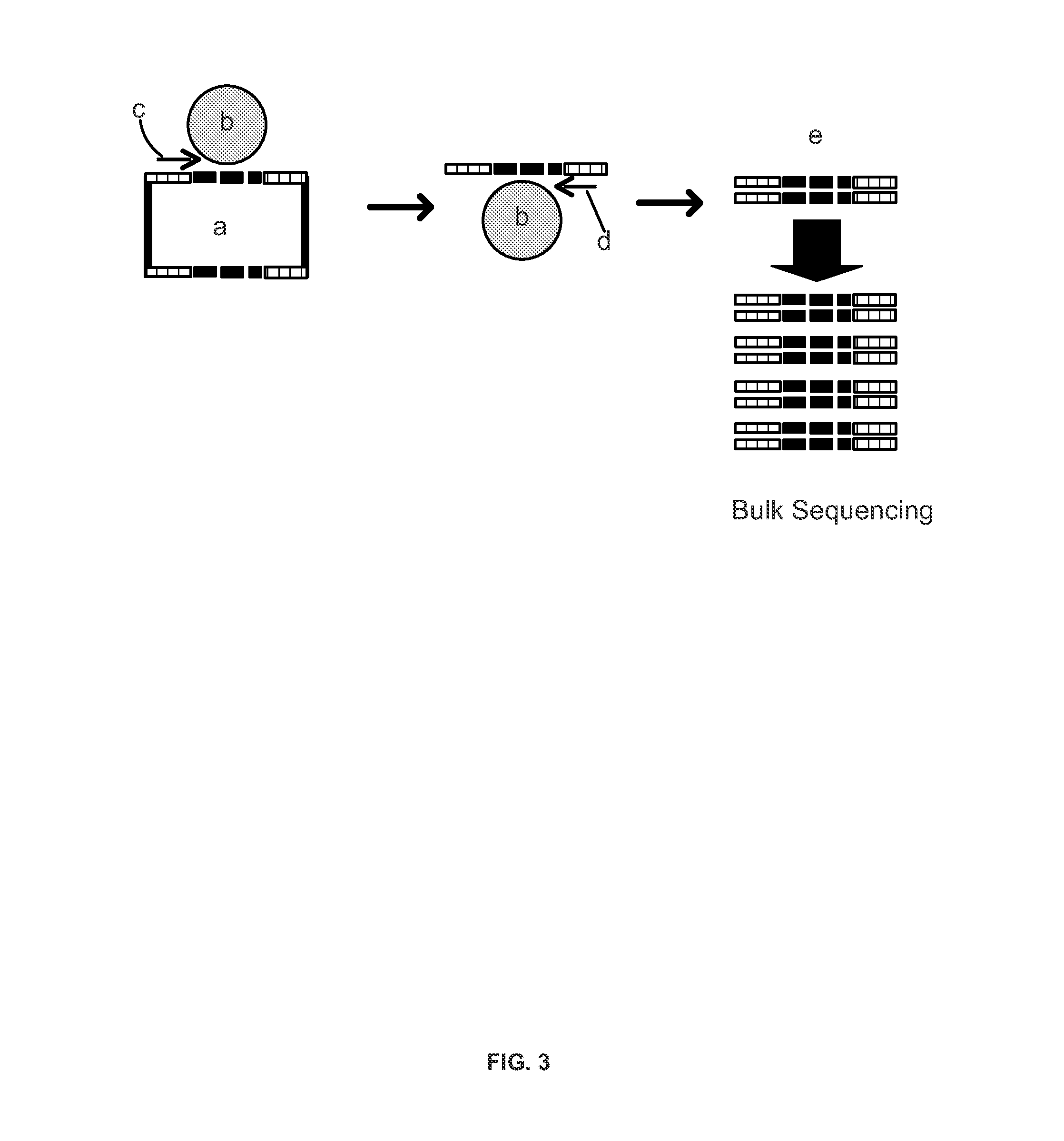
[0190]In one embodiment, a high-throughput protocol was implemented for human or mouse TCRβ repertoire library construction. The libraries were sequenced directly on the GAIIx next-gen sequencing platform (Illumina). For human samples, multiplex PCR was performed using a set of 20 primers to amplify across all 50 V segments and 10 primers to amplify across all 13 J segments. The primers libraries generated libraries that were the reverse complement of the native TCRβ sequence. This enabled sequencing from the J side of the constructs without further manipulation. The primers also had tails with the same sequence as a portion of the Illumina TruSeq library adapter. The 30 primers were pooled in a single 400 μl PCR, which contained genomic DNA from at least 5×105 cells. The reactions were then thermocycled for no more than 25 cycles, depending on the number of input cells. After thermocycling, a PCR column (Qiagen) was u...
example 3
Protocol Optimization Using 48-Plex Pool of TCRβ Plasmid Clones
[0192]The true content of any particular TCRβ repertoire is not known, so an endogenous TCRβ repertoire cannot serve as a gold standard for protocol optimization. A 48-plex pool of mouse TCRβ plasmid clones was designed to act as template for protocol optimization. First, multiplexed amplification was performed of the mouse TCRβ repertoire as described in Example 2. The PCR products were subcloned using the TOPO-TA vector (Life Technologies), transformed post ligation into TOP10 competent cells (Life Technologies), and 48 transformed colonies were picked. Next, the clones were sequenced by Sanger sequencing to identify the TCRβ clonotype sequences. All of the clones were unique, and represented a broad range of possible V-Jβ combinations. The plasmids were then mixed in a single tube, across three orders of magnitude and with six replicates at each concentration.
[0193]The 48-plex mixture was used to optimize the TCRβ amp...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com