NOVEL 7Beta-HYDROXYSTEROID DEHYDROGENASE MUTANTS AND PROCESS FOR THE PREPARATION OF URSODEOXYCHOLIC ACID
a technology of ursodeoxycholic acid and hydroxysteroid dehydrogenase, which is applied in the field of new 7beta-hydroxysteroid dehydrogenase mutants and process for the preparation of ursodeoxycholic acid, can solve the problems of incomplete reaction of educt and higher processing cos
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
production example 1
Identification of 7β-HSDH Activity
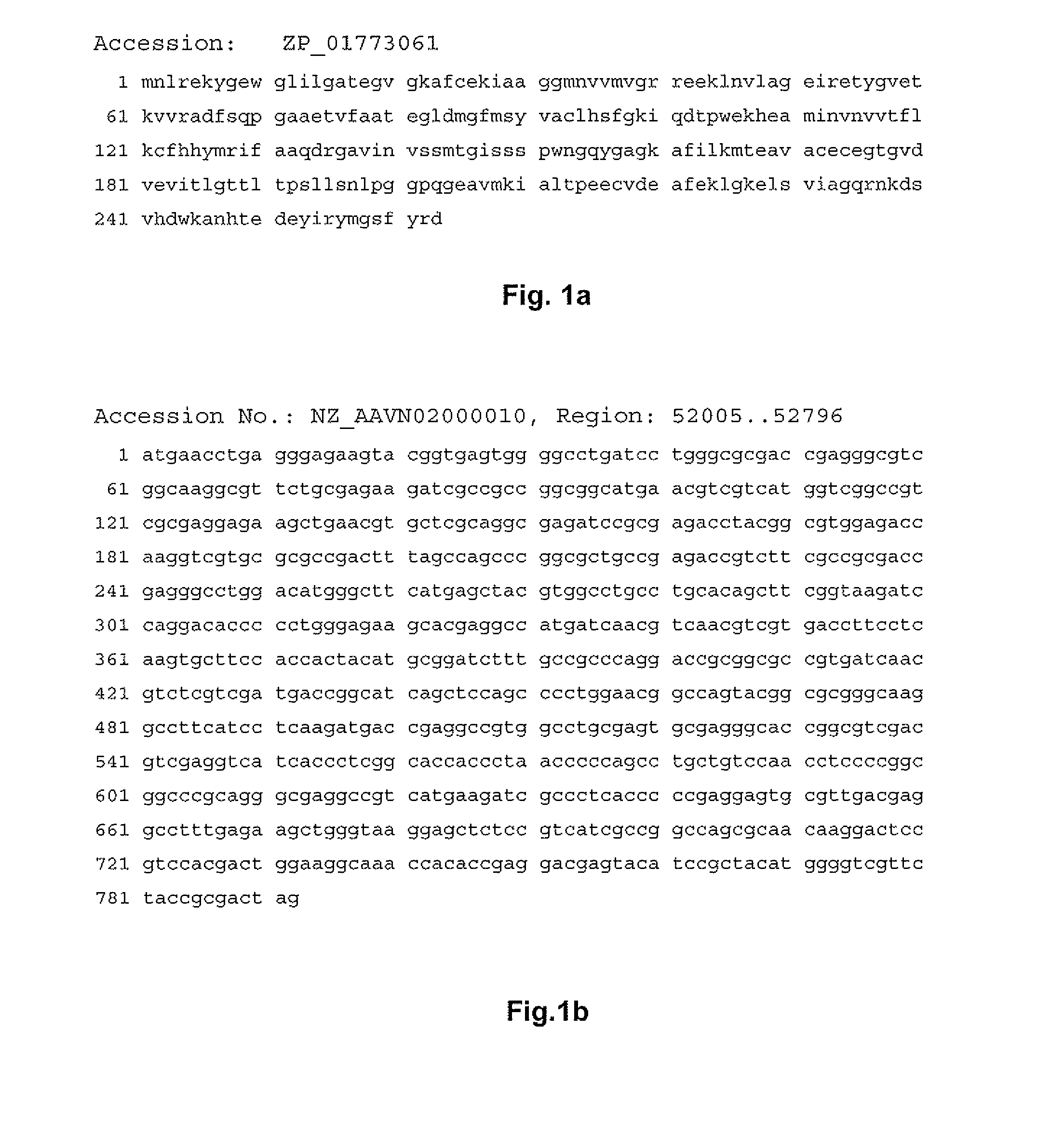
[0274]The genomic DNA sequence of Collinsella aerofaciens ATCC 25986 was published in the year 2007 by “Washington University Genome Sequencing Center” for the “human gut microbiome project” at GenBank. HSDHs belong to the “short-chain dehydrogenases”. As the biochemical function of the “short-chain dehydrogenases” from Collinsella aerofaciens ATCC 25986 had not been annotated in GenBank, 9 candidates were cloned into vector pET22b+, and then expressed in E. coli BL21(DE3).
[0275]For this, 7β-HSDH coding sequences were PCR-amplified. The PCR products were obtained using the genomic DNA of Collinsella aerofaciens ATCC 25986 (DSM 3979) as template and the primers 5′-gggaattcCATATGAACCTGAGGGAGAAGTA-3′ (SEQ ID NO:3) and 5′-cccAAGCTTCTAGTCGCGGTAGAACGA-3′ (SEQ ID NO:4). The NdeI and HindIII cleavage sites in the primer sequences are underlined. The PCR product was purified using the PCR-Purification-Kit (Qiagen) and then cut with the enzymes NdeI and HindI...
production example 2
Preparative-Scale Cloning, Expression and Purification Of 7β-HSDH from Collinsella aerofaciens ATCC 25986 and Further Characterization of the Enzyme
2.1 Cloning and Production of an Expression Construct
[0280]The gene coding for 7β-HSDH was once again amplified from the genomic DNA by PCR and using primers, as described above for production example 1:
[0281]The PCR product was once again purified as described above and digested with the restriction endonucleases NdeI and HindIII. The digested PCR product was purified again and cloned into the pET-28a(+) vector using the T4-ligase, to produce an expression vector. The resultant expression construct was then transformed into E. coli DH5α cells. The protein to be expected should have 20 amino acid residues comprising a signal peptide and an N-terminal 6×His-Tag and a thrombin cleavage site. The sequence of the inserted DNA was verified by sequencing.
2.2 Overexpression and Purification of 7β-HSDH
[0282]E. coli BL21(DE3) was transformed with...
production example 3
Production of 7β-HSDH Mutants and Characterization Thereof
[0295]Position 39 of the amino acid sequence (comprising start methionine) (cf. SEQ ID NO:2) was mutated.
3.1 Primers
[0296]The mutagenesis primers stated below were used for the site-directed mutagenesis of 7β-HSDH. The primers were selected based on the 7β-HSDH gene sequence, so that they bring about the desired amino acid exchange. It was borne in mind that the base to be mutated is localized centrally in the primer, and that the melting points of the primer pairs are in the same region.
[0297]The primer pair 7beta_mut_G39A_fwd and 7beta_mut_G39A rev was used for preparing the G39A mutant. The primer pair 7beta_mut_G39S_fwd and 7beta_mut_G39S_rev was used for preparing the G39S mutant.
Glycine → AlanineForward: 7beta_mut_G39A_fwd:CGTCGTCATGGTCGCCCGTCGCGAGG.SEQ ID NO: 9)Reverse: 7beta_mut_G39A_rev:CCTCGCGACGGGCGACCATGACGACG.SEQ ID NO: 10)Glycine → SerineForward: 7beta_mut_G39S_fwd:CGTCGTCATGGTCAGCCGTCGCGAGG.SEQ ID NO: 11)Revers...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com