Diagnostic methods for infectious disease using endogenous gene expression
a technology of endogenous gene expression and diagnostic methods, applied in biocide, instruments, biochemistry apparatus and processes, etc., can solve the problems of limited application and accuracy, inadequate white blood cell count,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0138]This example illustrates detection of viruses in young children with Fever Without an Apparent Source (FWS).
[0139]Subjects were drawn from as study of children between 2 to 36 months of age with Fever Without an Apparent Source (Table 1) and afebrile children having ambulatory surgery who were recruited at St. Louis Children's Hospital as described previously (Colvin, J. M., et al. 2012, Pediatrics 130(6):e1455-1462). The febrile and afebrile groups were similar with respect to age, gender, and season of recruitment, but differed with respect to race, with more African-American children in the febrile group (57% vs. 13%). Patients were enrolled according to Institutional Review Board—approved protocol. The study was approved by the Washington University Human Research Protection Office. Each subject was tested for multiple viruses in blood and nasopharyngeal samples using panels of virus-specific PCR assays as described (Colvin, J. M., et al. 2012, Pediatrics 130(6):e1455-1462...
example 2
[0140]This example illustrates preparation of biological samples, including RNA from blood samples.
[0141]In these investigations, whole blood and nasopharyngeal samples were collected for virus-specific PCR and high-throughput sequencing. Additionally, a blood sample was collected in a Tempus™ Blood RNA Tube (Applied Biosystems, Carlsbad, Calif.) and stored at −80° C. for subsequent gene expression analysis.
[0142]Total RNA was isolated from whole blood collected in Tempus™ Blood RNA tubes (Applied Biosystems, Carlsbad, Calif.) according to the manufacturer's instructions. RNA quality was determined by gel-chip image (showing 28S, 18S and 5S bands) and RNA integrity number (RIN, generally a>7 RIN indicates good quality RNA) using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, Calif.). All but 3 of the RNA preparations had RIN scores≧7.0. Total RNA concentration was obtained from an absorbance ratio at 260 nm and 280 nm using a NanoDrop ND-100 spectrometry instrument (NanoDrop Inc.,...
example 3
[0143]This example illustrates gene expression microarray assays.
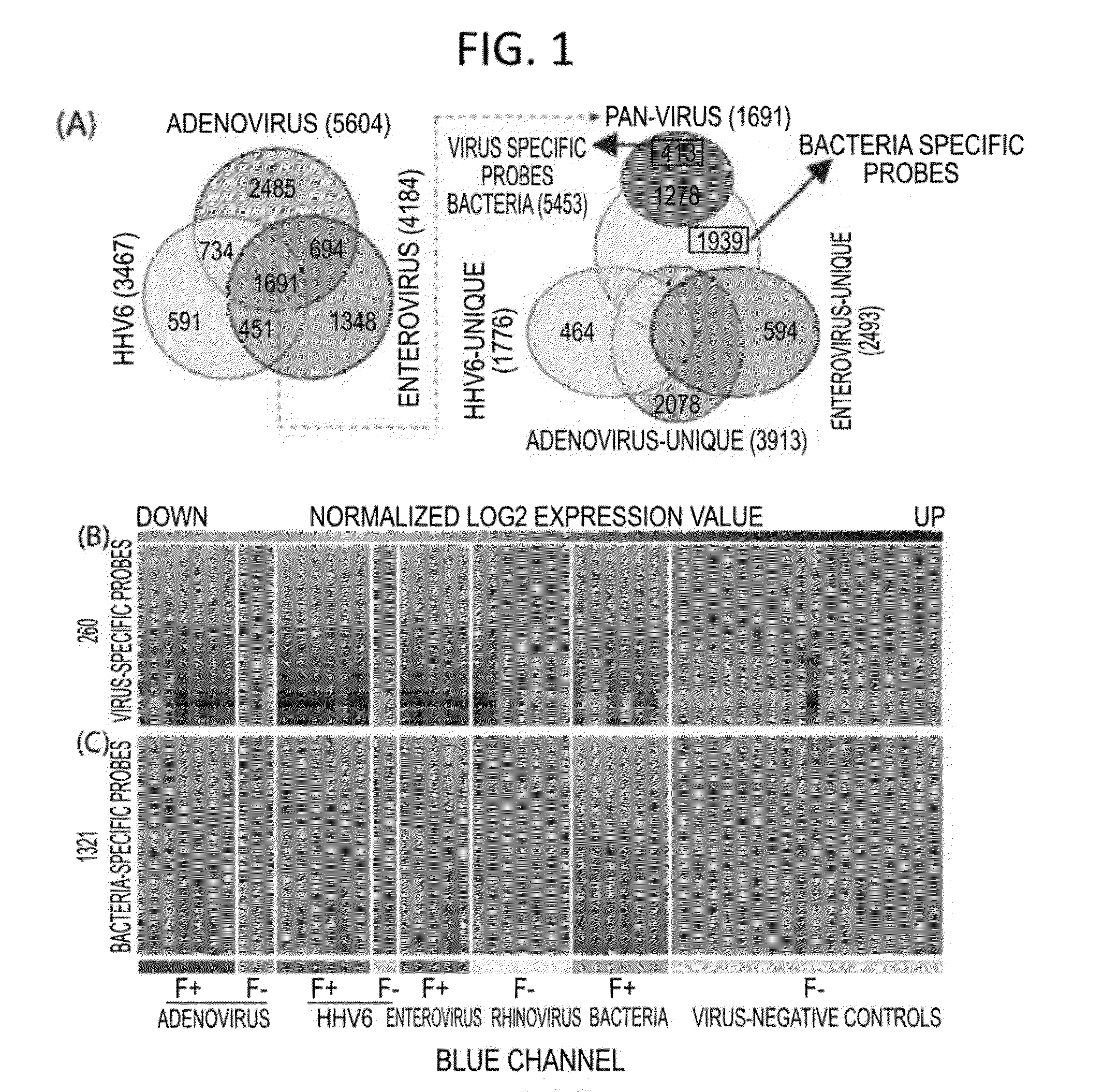
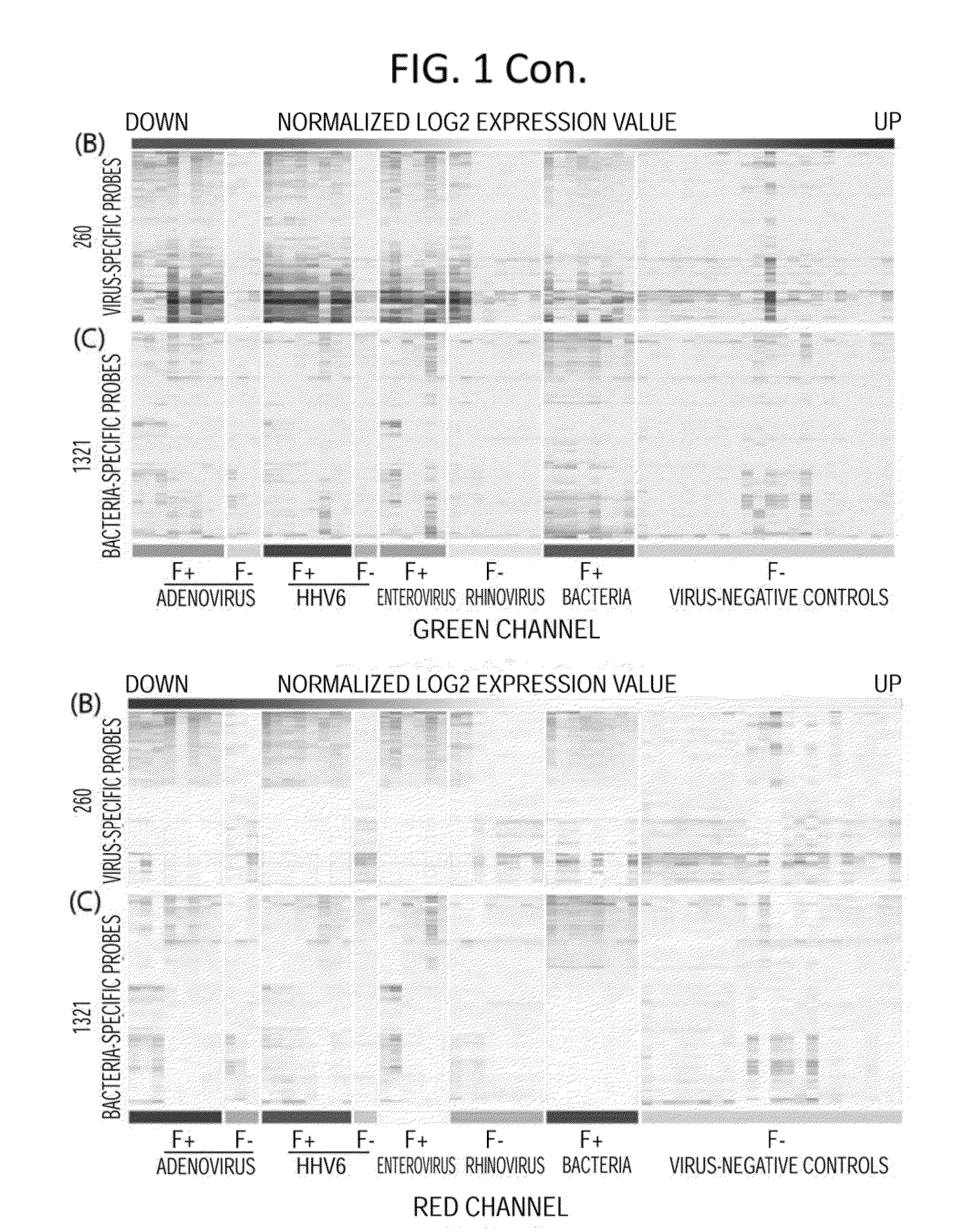
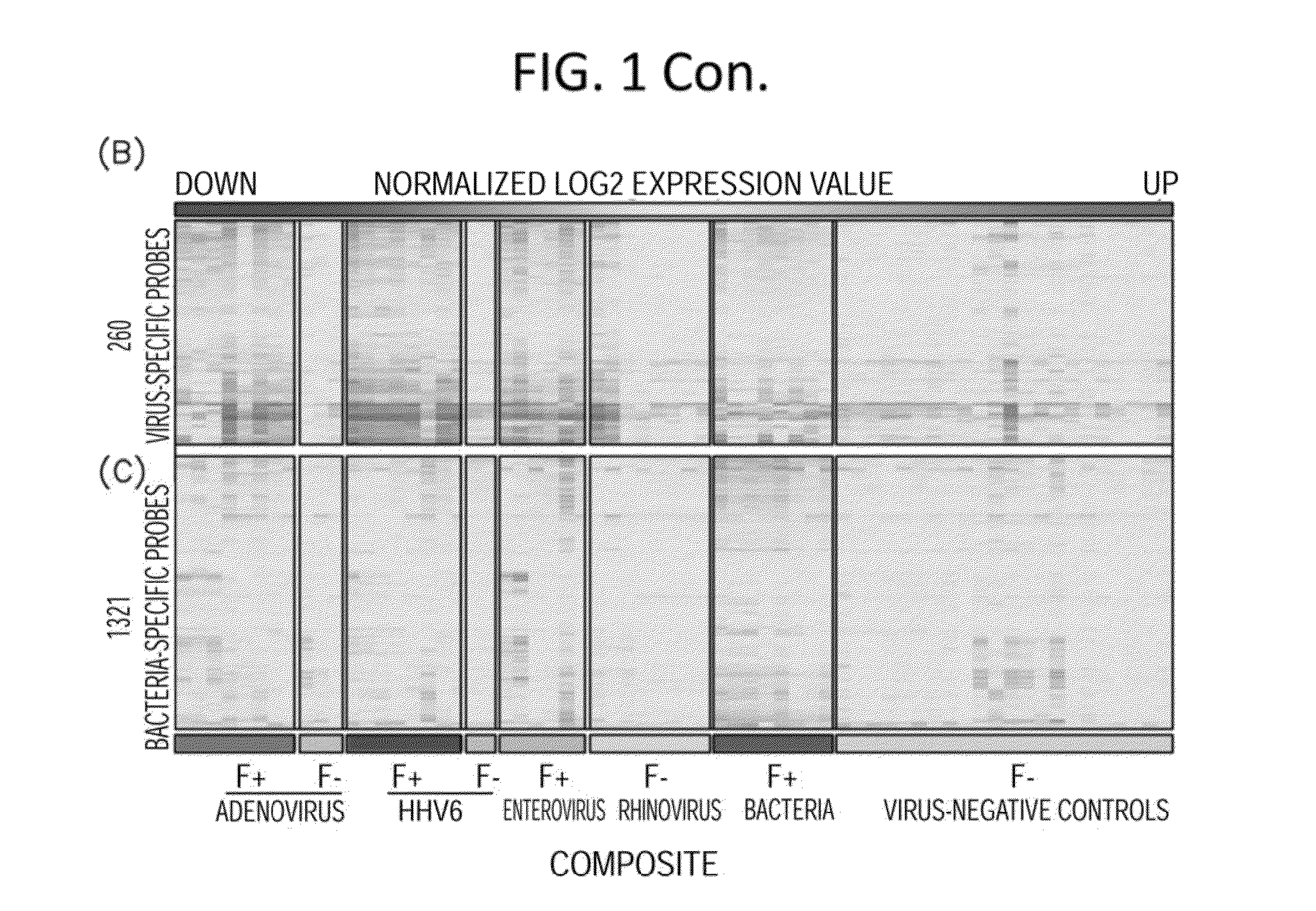
[0144]In these investigations, gene expression microarray analyses were conducted on blood samples from 35 febrile children positive for adenovirus, human herpesvirus 6 (HHV-6), or enterovirus infection or with acute bacterial infection, and 22 afebrile controls. Gene expression microarray assays were carried out at the Genome Technology Access Center in Washington University in St Louis. RNA transcripts were amplified by 17 linear amplification with the IIlumina 3TVT Direct Hybridization Assay Kit (Illumina Inc., San Diego, Calif.), and biotin-labeled cRNA targets were hybridized to the Illumina Human-HT12 v4 Expression BeadChips (>47,000 probes), which were scanned on an Illumina BeadArray Reader. Scanned images were quantitated by Illumina Beadscan software (v3). Quantified data were imported into Illumina GenomeStudio software (version 2011.1) to generate expression profiles and to make data quality assessments. Th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| Northern blot assay | aaaaa | aaaaa |
| dual color fluorescently | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com