Synthesis of 2', 3'-dideoxynucleosides for automated DNA synthesis and pyrophosphorolysis activated polymerization
a technology of dna and synthesis method, applied in the field of synthesis of various 2′, 3′-dideoxynucleoside supports, can solve the problems of inability to commercially synthesize dda, ddg, dt, and all such synthetic methods are labor-intensive and costly, and no adequate method for making dna containing dt has been described
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
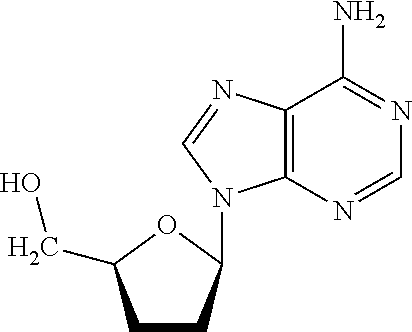
Preparation of A 2′,3′-Dideoxyadenosine Support Structure
[0098]
[0099]9-(5-((tris(4-methoxyphenyl)methoxy)methyl)tetrahydrofuran-2-yl)-9-purin-6-amine (product 12 in Reaction Scheme H, above) was synthesized first according to the reaction shown by Reaction Scheme H. To obtain this compound, 2′,3′-dideoxyadenosine (500 mg, 2.13 mmol) was dried three times by co-evaporation with pyridine, and suspended in about 10 mL of dry pyridine, were added DMTC1 (1.084 g, 3.2 mmol), triethylamine (0.45 mL, 3.2 mmol) and DMAP 6.48 mg, 0.053 mmol) were added. This reaction mixture was stirred at room temperature for about 4 hours, and then about 10 mL of 5% NaHCO3 were added.
[0100]The mixture was then extracted with two ˜30 mL portions of ethyl acetate, followed by combining the organic layers and by evaporation to dryness. The residue was purified by flash chromatography on silica gel (CH2Cl2 / CH3OH=10 / 1), to create product 12, i.e., 9-(5-((tris(4-methoxyphenyl)methoxy)methyl)tetrahydrofuran-2-yl)-...
example 2
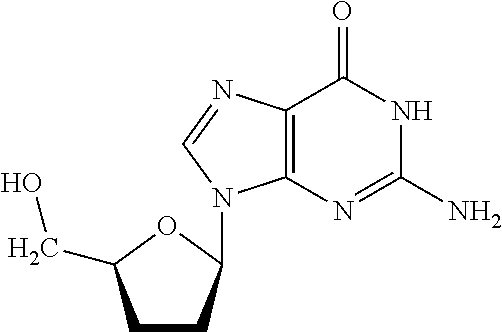
Preparation of A 2′,3′-Dideoxyguanosine Support Structure
[0104]
[0105]9-(5-(hydroxymethyl)tetrahydrofuran-2-yl)-6-oxo-6,9-dihydropurin-2-yl)-N,N-dimethylformimidamide amine (product 3 in Reaction Scheme A, above) was synthesized first according to the reaction shown by Reaction Scheme A. To obtain this compound, 2′,3′-dideoxyguanosine (about 947 mg, 3.77 mmol) was dissolved in 20 mL of methanol and N,N-dimethylformamide dimethyl acetal (about 1.06 mL, 7.98 mmol) was added. The mixture was stirred overnight at room temperature followed by adding another portion of N,N-dimethylformamide dimethyl acetal (about 0.5 mL, 3.9 mmol). The mixture was again stirred for about 2 hours at room temperature. The solvent was evaporated to yield the product, (compound 3 in Reaction Scheme A, above) as a white solid.
[0106]Next, compound 3 as shown in reaction scheme A was used to obtain N,N-dimethyl-N′-(6-oxo-9-(5-((tris(4-methoxyphenyl) methoxy)methyl)tetrahydrofuran-2-yl)-6,9-dihydropurin-2-yl) form...
example 3
Preparation of N-(9-(5-(1-(4-methoxyphenyl)-1-phenylethoxy)methyl) tetrahydrofuran-2-yl)-6-oxo-6,9-dihydro-1H-purin-2-yl)isobutyramide
[0111]
[0112]Compound 5A shown in Reaction Schemes E and F, above, can be used for preparing compound 6 shown in Reaction Schemes C and F, instead of compound 5 discussed in Example 2, above. The synthesis was carried out according to the reaction showin by Reaction Scheme D. 2′,3′-dideoxyguanosine was co-evaporated with anhydrous pyridine three times before it was suspended in about 20 mL of dry pyridine. Trimethylchlorosilane (about 1.6 mL, 12.4 mmol) was added and the reaction mixture was stirred at room temperature for about 15 minutes, followed by adding about 2.6 mL (15.5 mmol) isobutyric anhydride. The reaction mixture was kept overnight at room temperature, followed by cooling in an ice bath and adding about 4 mL of water after about 5 minutes. Then, about 4 mL of about 29% aqueous ammonia was added and the reaction was stirred for about 15 min...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com