Method for detecting opioids, opiates, cannabinoids, or benzodiazepines in a sample with a b-glucuronidase enzyme
a technology of b-glucuronidase and enzyme, applied in the field of biotechnology, can solve the problems of corroding vulnerable equipment, stricter security conditions, and degrading sensitive compounds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
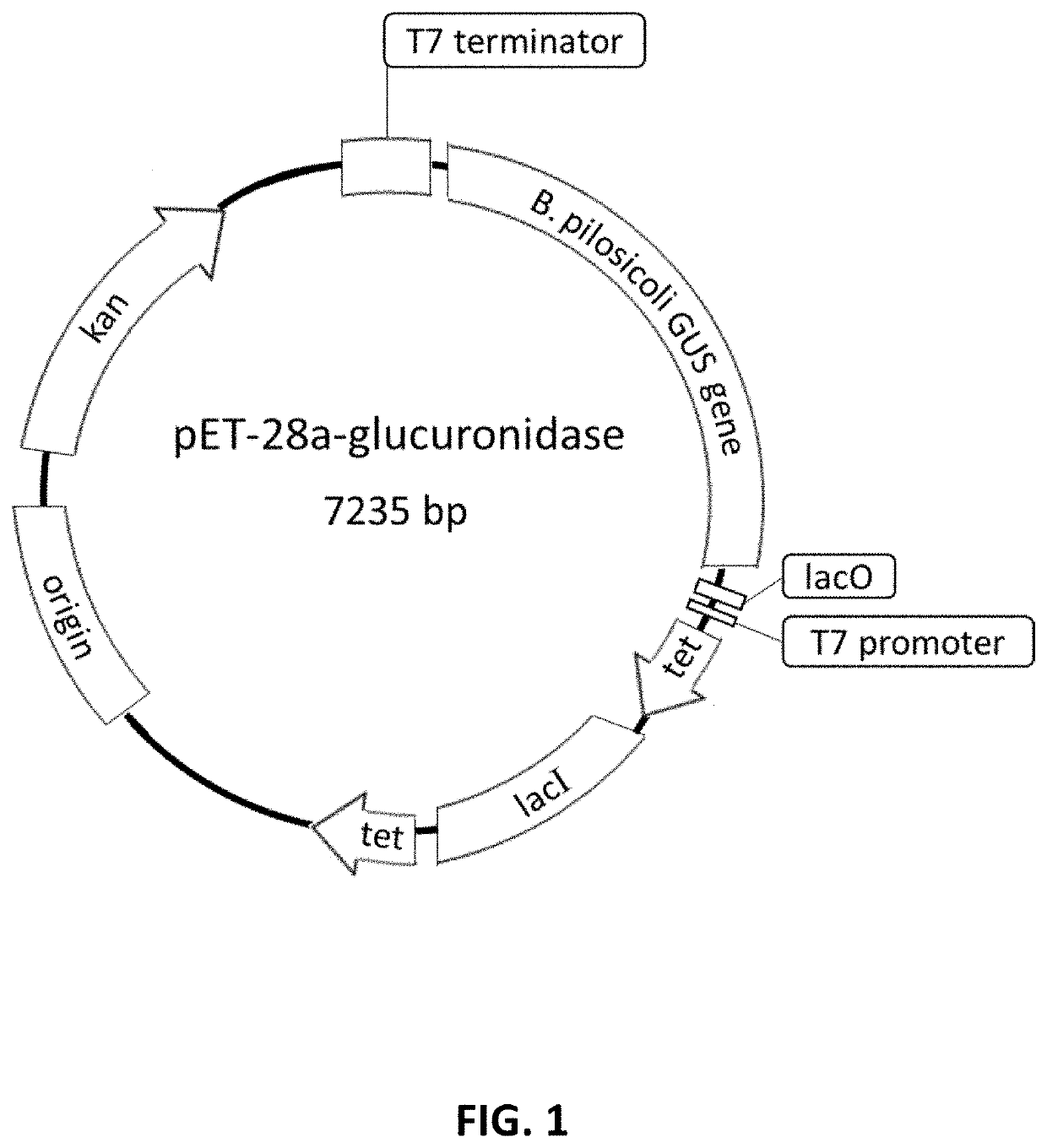
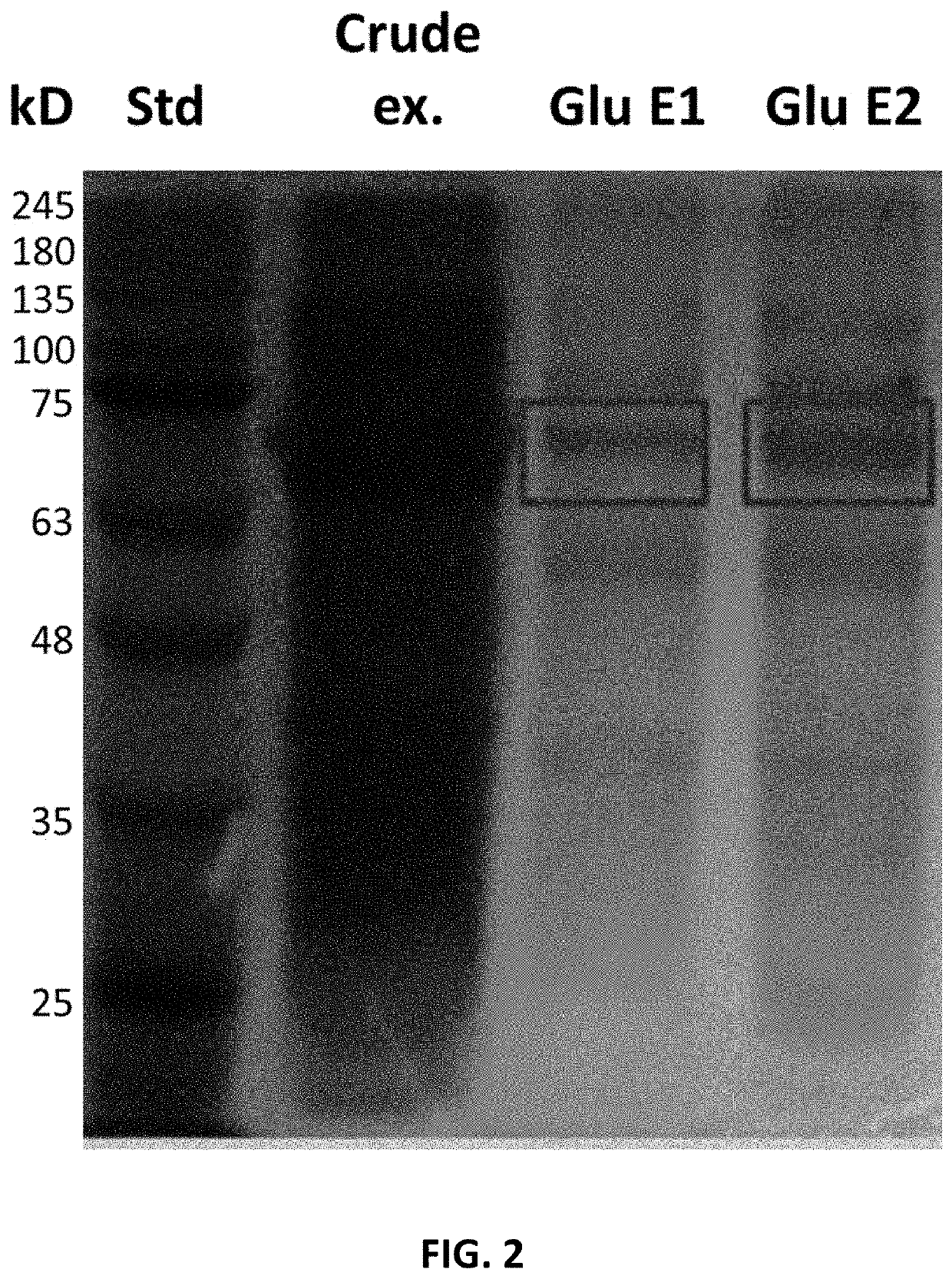
and Expression of β-glucuronidase Enzyme from B. pilosicoli
[0045]To express the β-glucuronidase enzyme, nucleotide sequence from said protein was used, originated from Brachyspira pilosicoli strain B2904 described in Genbank (access code CP003490), which is shown in SEC.ID.NO.2, and codon optimization was performed using the algorithm OptimumGene™ (GenScript) to efficiently express said sequence in E. coli. To do this, codon adaptation index (CAI), understood as distribution of frequency of codon usage through the sequence, was adjusted from 0.66 to 0.88, in which a CAI value of 1.0 is considered to be perfect in the desired expression organism; and a CAI value greater than 0.8 is regarded as good in terms of high gene expression levels. Guanine-cytosine content was also optimized to extend mRNA half-life and stem-loop structures were eliminated, since they disrupt ribosome binding and mRNA stability. Optimized nucleotide sequence obtained is shown in SEC.ID.NO.3. Amino acid sequen...
example 2
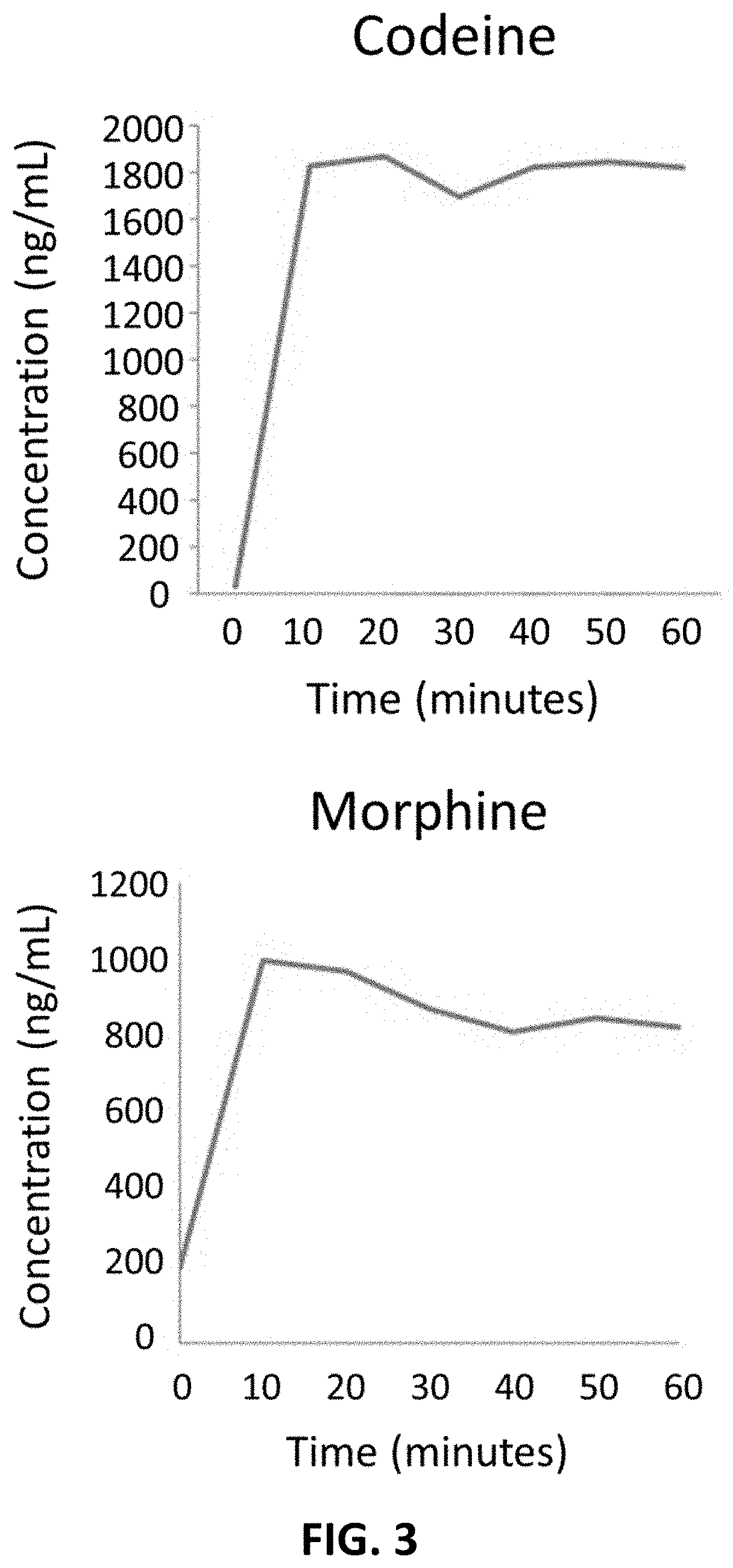
n Time of a Sample with β-glucuronidase from B. pilosicoli
[0050]The required time to hydrolyze morphine-3-glucuronide and codeine-6-glucuronide with the enzyme β-glucuronidase in excess (named BGTurbo™) from B. pilosicoli was evaluated with the following protocol: 10 μL of glucuronide metabolites at a concentration of 100 μg / mL, free standards (free codeine and morphine calibrators at concentrations of 312.5, 625, 1,000, 1,250 and 1,875 ng / mL), and internal standards (40 μL of deuterated free drug at a concentration of 1 μg / mL to each sample) were added to a sample of 0.4 mL of blank urine and then submitted to vortex agitation. Then, 360 μL of sodium phosphate (pH 7; 140 mM) solution was added to each sample. Then, 240 μL of β-glucuronidase (BGTurbo™) were added to each sample and then agitated by vortex. Samples were incubated at 55 degrees Celsius in an oven during 0, 10, 20, 30, 40, 50 and 60 minutes. A total of 21 samples were used: 7 points of measure at different times, each...
example 3
on of Specific Activity of β-glucuronidase from B. pilosicoli
[0060]Specific activity of the β-glucuronidase enzyme from B. pilosicoli (BGTurbo™) was calculated and compared with a mean value of three calculations of the specific activity of β-glucuronidase from E. coli, using the following protocol: 350 μL of sodium phosphate buffer solution (NaH2PO4) 0.1 M, pH 6.8 were mixed with 350 μL of phenolphthalein β-D-glucuronide sodium salt (0.64 mg / mL) and the mixture was incubated at 37 degrees Celsius. Then, 50 μL of solution containing the enzyme were added to the mixture and incubated at 37 degrees Celsius during 30 minutes. After the first 15 minutes the mixture was gently agitated and stopped at 30 minutes from the beginning of the reaction with 2.5 mL of a glycine solution 0.2 M, pH 10.4. Absorbance was read at 540 nm and the amount of enzyme used was calculated (U / mL) when adjusting the values in a calibration curve. Protein concentration was measured using Bradford method, as wi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com