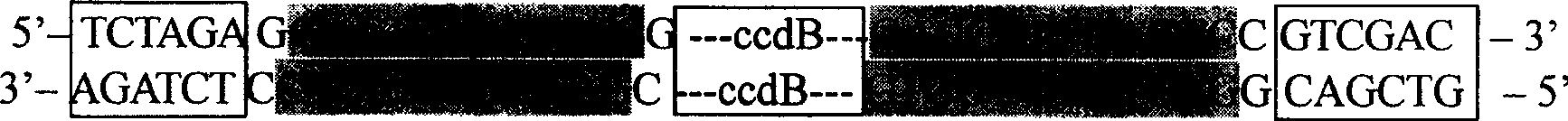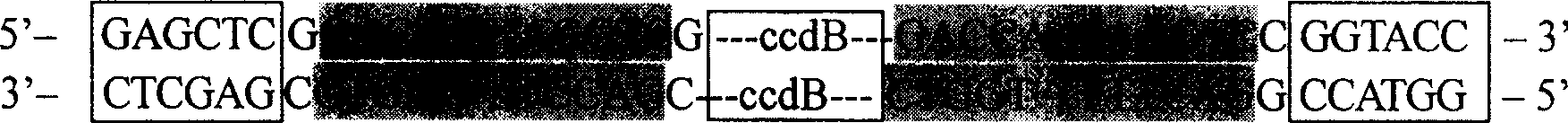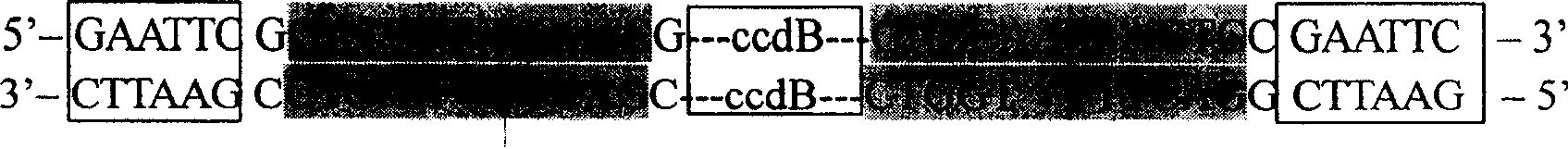A method of constructing T carriers
A carrier and enzyme cutting site technology, applied in the field of genetic engineering, can solve the problem of high background of non-recombinant transformants, and achieve the effect of improving quality and stability and excellent product characteristics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1, Construction of pSKΔAhd
[0042] Use the method of site-directed mutagenesis to silence the AhdI restriction site of the Amp resistance gene in the pBlueScript SK(-) vector to obtain pSKΔAhd. ) vector as a template, PCR amplification was carried out with Pfu DNA polymerase. Among them, the PCR amplification primers are (boxed C is the introduced mutation site (G in the original sequence), and the shaded part is the AhdI restriction site after mutation destruction):
[0043] AmpF: 5'-CTACGATACGGGAGGGCTTACCATCTGG-3';
[0044] AmpR: 5'-TTATCTACAC AGGCAAC-3';
[0045] The reaction system of PCR amplification is: 5U Pfu, 0.2mmol / L dNTPs, 1×Pfu buffer in 50μL reaction system, 10μmol / L of primers, about 5ng of template; the cycle program of PCR amplification is: 94℃ pre-denaturation for 5min , (94°C, 30S; 60°C, 30S; 72°C, 5min) × 30, 72°C for a final extension of 5min. After 1% agarose gel electrophoresis, the gel was cut to recover the PCR amplification produ...
Embodiment 2
[0049] Example 2. Introducing the AhdI box into the multiple cloning site of pSKΔAhd to construct the pre-T vector
[0050] Using the plasmid pENTR1A (purchased from Invitrogen) as a template, PCR amplification was performed with CcdB01F and CcdB01R primers. The reaction system of PCR amplification is: 5U Pfu, 0.2mmol / L dNTPs, 1×Pfu buffer in 50μL reaction system, 10μmol / L of primers, about 5ng of template; the cycle program of PCR amplification is: 94℃ pre-denaturation for 5min , (94°C, 30S; 60°C, 30S; 72°C, 1.5min) × 30, 72°C for a final extension of 5min. The PCR products were recovered by 1% agarose gel electrophoresis, and the recovered PCR product and pSKΔAhd plasmid were digested with SalI and XbaI respectively, and the digested products were recovered by 1% agarose gel electrophoresis. Establish the following ligation reaction system between the digested PCR product and the digested pSKΔAhd vector: 10 μL ligation system containing 3U ligase (Promega), 1× ligation buff...
Embodiment 3
[0068] Example 3, Preparation of pSKxx-T Series T Vectors and Analysis of TA Cloning Efficiency
[0069] 1. Preparation of T carrier
[0070] The pre-T vectors pSK01, 02, 03 and 04 were respectively digested with AhdI, and the enzyme digestion reaction system was as follows: 20 μL digestion system contained 3U AhdI endonuclease (purchased from NEB Company), 1×NEBuffer4, and about 1 μg plasmid. After enzyme digestion at 37°C for 3 hours, large fragments were recovered by 1% agarose gel electrophoresis. The large fragment recovered and purified by gel cutting is the corresponding T vector. The physical maps of pSK01-T, pSK02-T, pSK03-T and pSK04-T are as follows figure 2 , image 3 , Figure 4 with Figure 5 shown.
[0071] 2. Test TA cloning efficiency
[0072] Arabidopsis NCED3 (1.8kb, GenBank Accession No.AT3G14440) and DREB1A (680bp, GenBank Accession No.At4g25480) were amplified by PCR using Taq DNA polymerase with NCEDF and NCEDR as primers, respectively fragment. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com