Novel expression enzyme yeast gene engineering system
A technology of genetically engineered bacteria and genes, applied in the field of yeast genetic engineering systems, can solve the problems of no product development, no work report, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
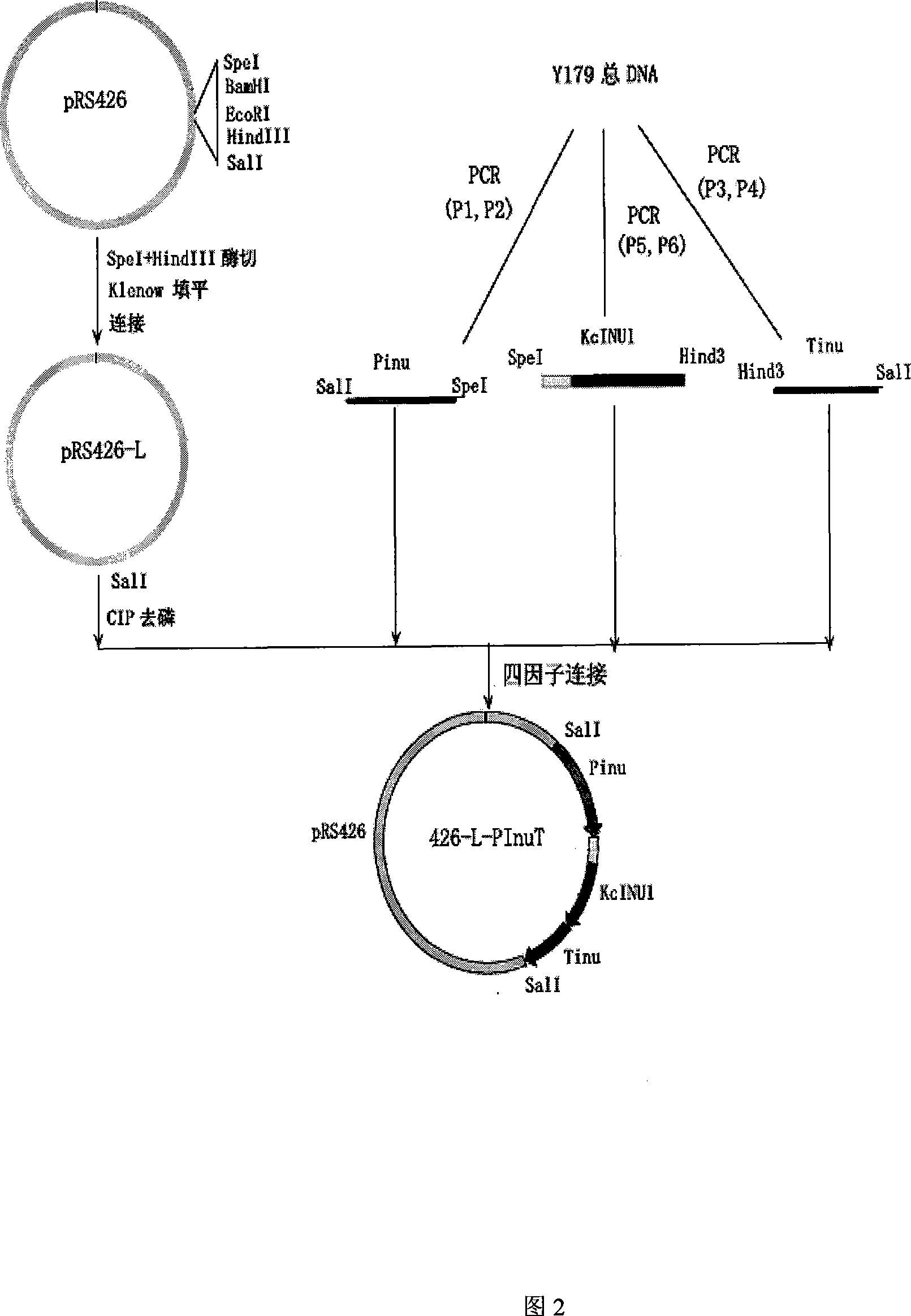
[0048] Example 1 Construction of Kluyveromyces chickpea inulinase gene expression cassette
[0049] First, using the total DNA of Kluyveromyces chickpea Y179ura3- as a template, using P1 and P2, P3 and P4, and P5 and P6 as primers, the promoter and terminator of the inulinase gene were amplified by PCR and the DNA fragment sequenced with the inulinase gene sequence with the signal peptide sequence, and the corresponding restriction endonuclease treatment was carried out according to the enzyme cutting site designed for each fragment, and then these three fragments were combined with the plasmid pRS426 digested by Sal1 -L four factors were connected to obtain plasmid 426-L-PInuT, which included the promoter of the inulinase gene, the terminator and the Sal1 fragment sequenced by the inulinase gene with the signal peptide sequence, which was spliced Inulinase gene expression cassette. Among them, pRS426-L is derived from pRS426, and the structure of pRS426 can be found in: Siko...
Embodiment 2
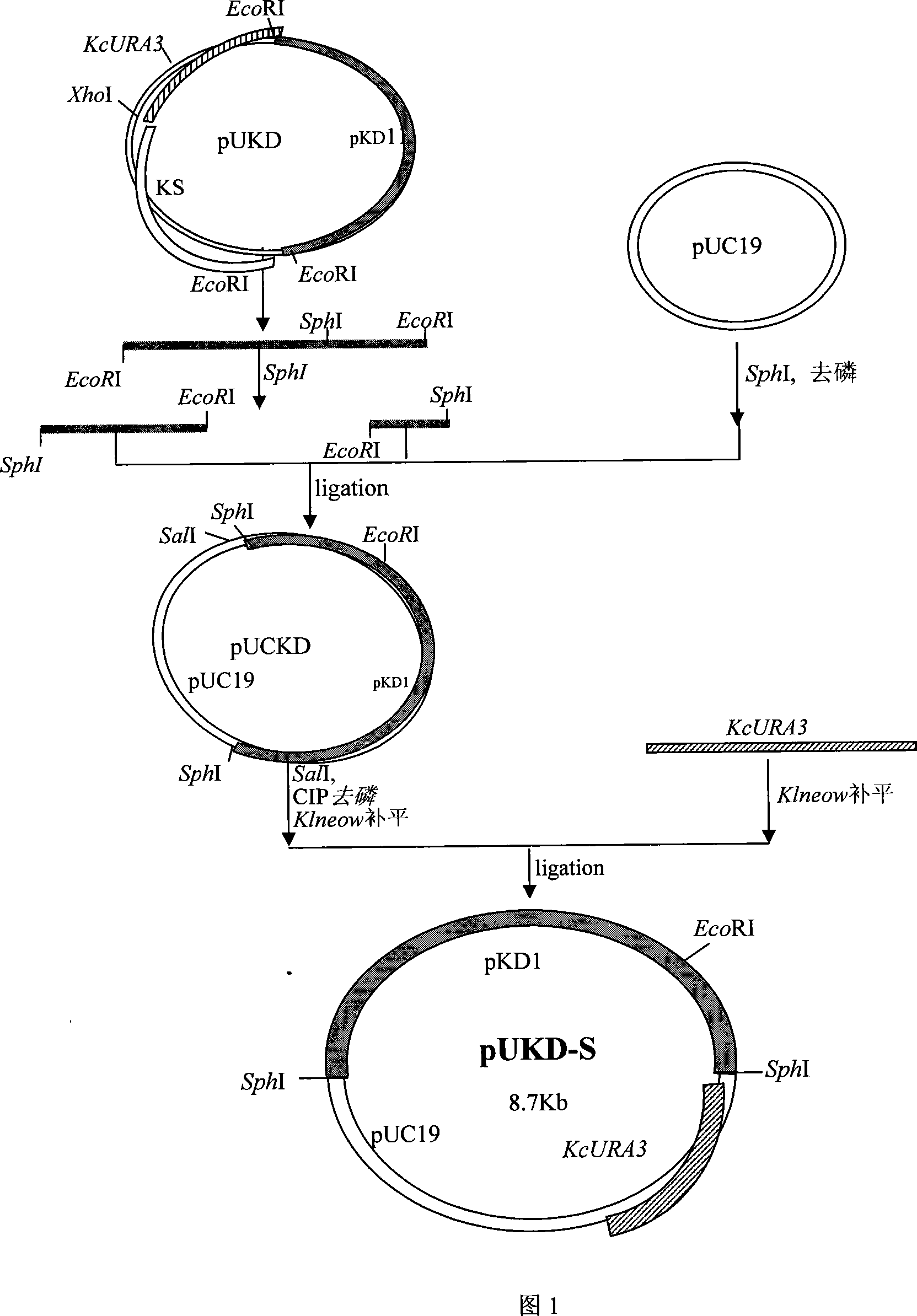
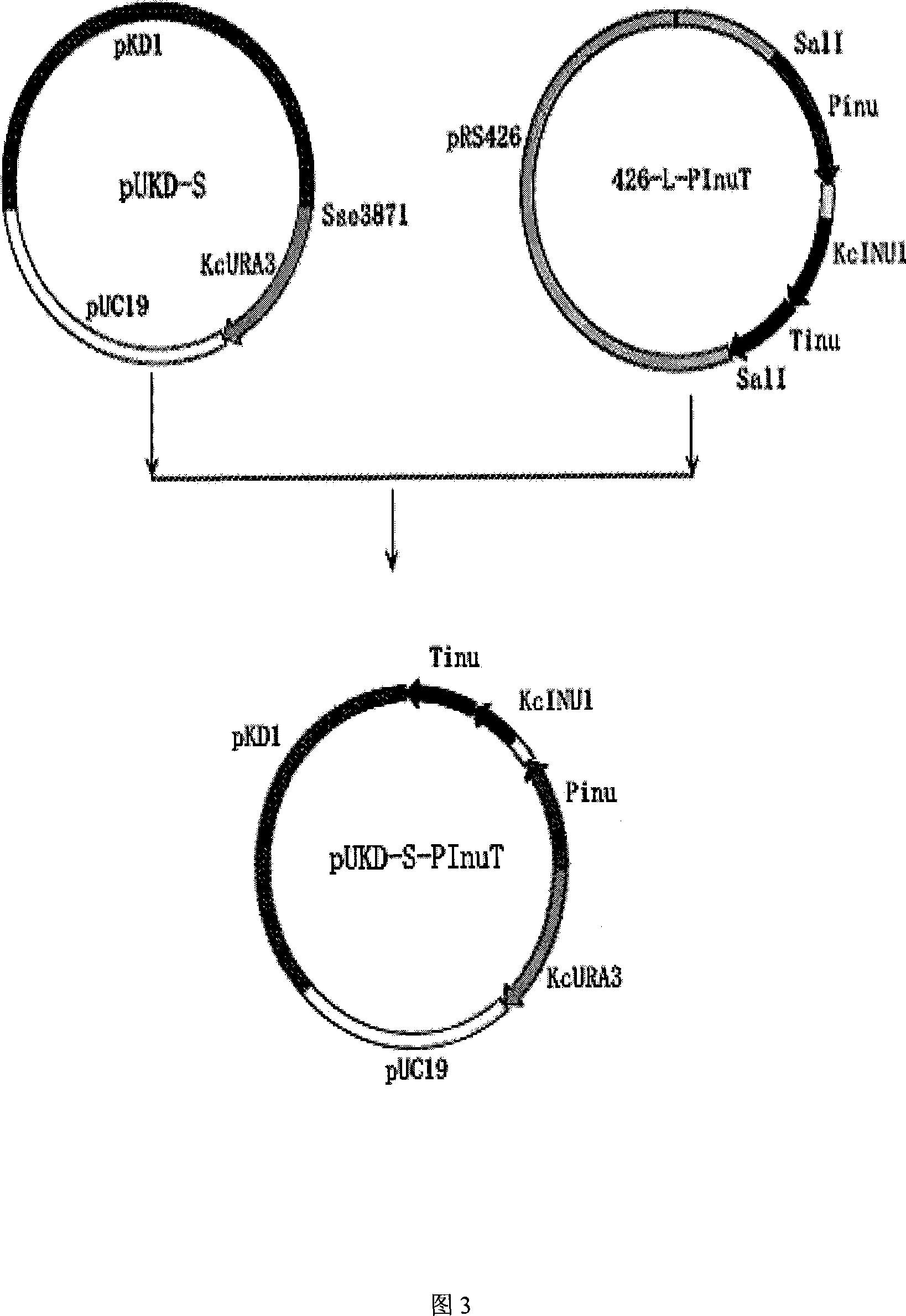
[0065] Example 2 Construction of Kluyveromyces chickpea inulinase gene expression plasmid pUKD-S-PInuT
[0066] First, the plasmid 426-L-PIT-T in Example 1 was excised with the restriction endonuclease Sall to cut out the DNA fragment with the inulinase gene expression cassette. After the fragments were separated, the sticky ends were filled in with Klenow enzyme; the Kluyveromyces chickpea vector pUKD-S was cut with restriction endonuclease Sse3871, and the sticky ends were also filled in with Klenow enzyme. Then, the two are connected with DNA ligase to obtain the Kluyveromyces chickpea inulinase gene expression plasmid pUKD-S-PInuT.
Embodiment 3
[0067] Example 3 Inulinase Gene Expression Plasmid pUKD-S-PInuT Using LiAc Complete Cell Transformation Method
[0068] ① Inoculate a single colony into 3ml YEPD and culture overnight at 30°C with shaking.
[0069] ② Dilute the overnight culture into 50ml YEPD (10 tubes can be made) so that the initial concentration is O.D 600 =0.08, shake culture at 30℃ for 4.5h to O.D 600 = 0.32. (Initial concentration and incubation time have a great influence on the transformation efficiency.)
[0070] ③5K, 5min centrifugation to collect bacteria
[0071] ④ Discard the culture medium, add sterile water at 5K, and collect the bacteria by centrifugation for 5 minutes.
[0072] ⑤ Wash once more with sterile water, centrifuge at 5K for 5 minutes to collect bacteria.
[0073]⑥ Add 10ml of 0.1M LiAC, incubate in a 30°C water bath for 10min, and divide the cells into 1.5ml centrifuge tubes, 1ml per tube.
[0074] ⑦Centrifuge the cells at high speed and discard the supernatant.
[0075] ⑧ A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com