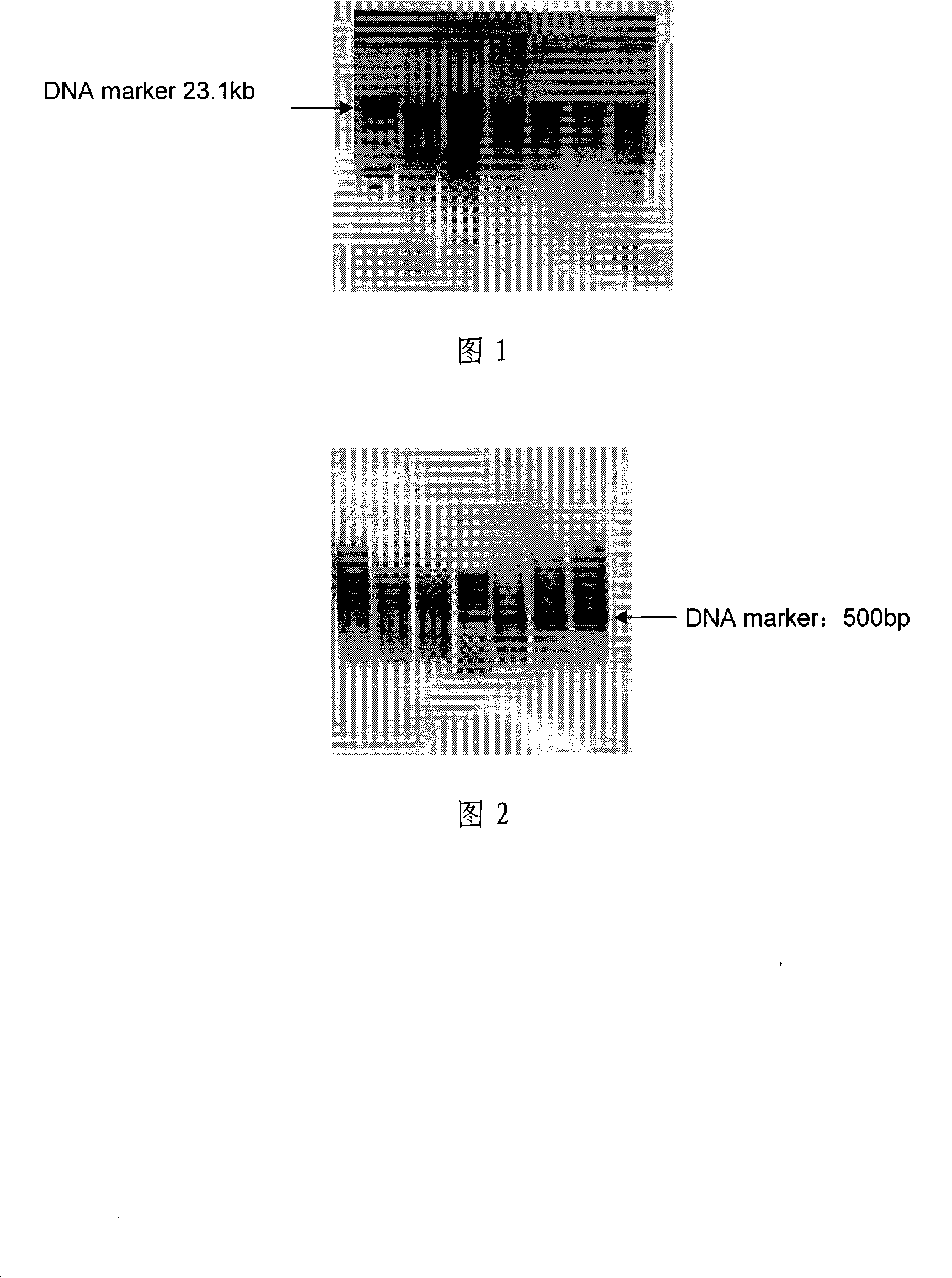
Soil sample total DNA extraction method for improving DNA quality
A soil sample and extraction method technology, applied in the field of soil microbiology, can solve the problems of limited removal capacity and large difference in DNA quality, and achieve the effects of small cost increase, easy PCR amplification process and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] 1) Accurately weigh 0.5g of northeast black soil soil sample, place it in a 2mL sterilized centrifuge tube, add 0.3g of quartz sand, and then add 800-1000μL of DNA extraction buffer (100mM Tris-HCl[pH8.0], 100mM EDTA [pH8.0], 100 mM sodium phosphate [pH8.0], 1.5M NaCl, 1% CTAB) and 5 μL proteinase K (20 mg / mL). Vortex and vibrate for 10 minutes to fully disperse the soil aggregate structure and fully mix with DNA extraction buffer and other components. Place in a 37°C water bath for 1 hour. During the water bath process, turn the sample over and mix several times every 15 minutes; the phosphate buffer It is 0.147g sodium dihydrogen phosphate; 5.08g disodium hydrogen phosphate heptahydrate can be dissolved in 200 ml of water.
[0022] 2) Add 200 μL of 10% SDS solution to the reaction solution after shaking, vortex to mix, and then freeze and thaw repeatedly 3 times. The freezing time at ℃ is 30 minutes, and the melting time at 65 ℃ is 20 minutes. Then put it in a 65°C ...
Embodiment 2
[0030]1) Accurately weigh 1.0g of Changbai Mountain Korean pine forest soil sample, place it in a 5mL sterilized centrifuge tube, add 0.5g of quartz sand, and then add 1500μL of DNA extraction buffer (100mM Tris-HCl[pH7.5], 100mM sodium EDTA [pH7.5], 100 mM sodium phosphate [pH7.5], 1.5M NaCl, 1% CTAB) and 15 μL proteinase K (20 mg / mL). Vortex and vibrate for 20 minutes to fully disperse the soil aggregate structure and fully mix it with DNA extraction buffer and other components. Place it in a 37°C water bath for 1.5 hours. The solution is 0.735g of sodium dihydrogen phosphate and 25.4g of disodium hydrogen phosphate heptahydrate dissolved in 1000 ml of water.
[0031] 2) Add 400 μL of 10% SDS solution to the reaction solution after shaking, vortex to mix, and then freeze and thaw repeatedly 3 times. 40min, melting time at 65°C is 10min. . Then put it in a 65°C water bath for 2 hours, and gently shake it every 15-20 minutes to fully lyse the cell wall; then centrifuge at 1...
Embodiment 3
[0039] The difference from Example 1 is:
[0040] 1) Accurately weigh 5g of soil sample, add 1g of quartz sand, and then add 5mL of DNA extraction buffer (50mM Tris-HCl[pH7.5], 80mM sodium EDTA[pH7.5], 60mM phosphate buffer (sodium phosphate) [pH7.5], 2.5M NaCl, 2% CTAB) and 50 μL proteinase K (20 mg / mL). Vortex and vibrate for 15 minutes to fully disperse the soil aggregate structure and fully mix with DNA extraction buffer and other components, place in a 30°C water bath for 2 hours, and turn the sample over and mix several times every 5 minutes during the water bath;
[0041] 2) Add 2000 μL of 10% SDS solution to the reaction solution after shaking, centrifuge at 8000×g for 30 min at room temperature, and collect the supernatant in a new sterilized centrifuge tube.
[0042] Add 600 μL of DNA extraction buffer and 40 μL of 10% SDS solution to the obtained precipitate, re-shock, dissolve the precipitate, and place it in a 65°C water bath for 50-60 min, centrifuge at 8000×g f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com