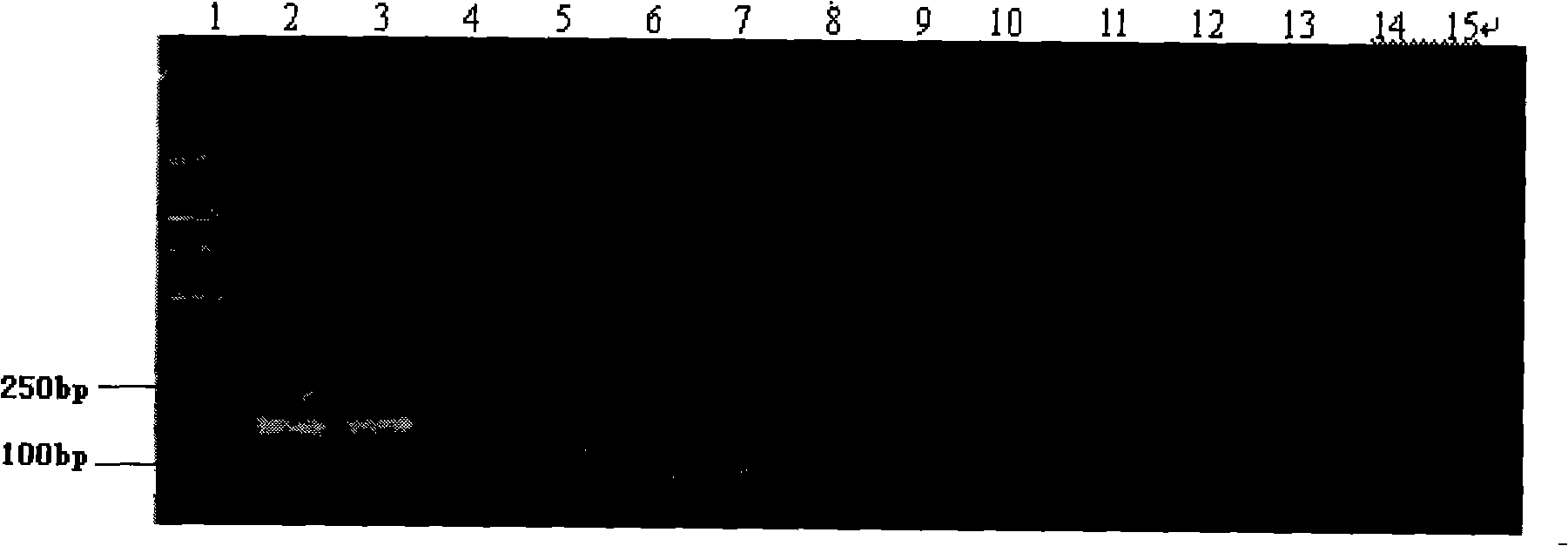Amplification primer for detecting pigeon source component by PCR-mtDNA, detecting kit and using method thereof
A technology of source components and amplification primers, applied in biochemical equipment and methods, recombinant DNA technology, DNA/RNA fragments, etc. The effect of spreading and spreading
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Detection and verification experiments on pigeons; feed containing components derived from pigeons; quail; chickens; ducks; geese; ostriches; partridges; cattle; sheep; horses; donkeys; fish animals.
[0041] (1) Extraction of total DNA:
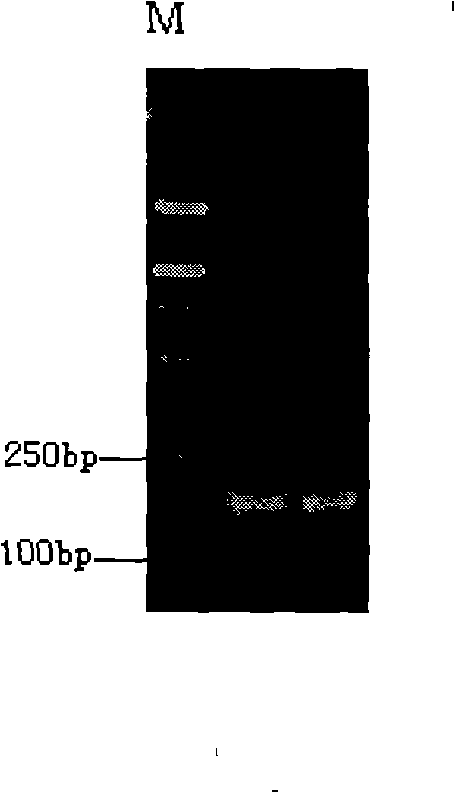
[0042] Using phenol chloroform extraction method to extract pigeon; feed containing pigeon-derived ingredients; quail; chicken; duck; goose; ostrich; partridge; cattle; sheep; horse; donkey; figure 1 . The purity and concentration of extracted genomic DNA were determined by UV spectrophotometer. The measured OD260 / OD280 values were both about 1.8, indicating that the DNA purity was high and met the requirements of PCR amplification.
[0043] (2) Design of PCR-mtDNA specific primers:
[0044] Compare the mitochondrial gene sequences of various animals published in GenBank, and design PCR-specific primers to identify and detect pigeon-derived components based on their species-conserved sequences (only specific fragments ...
Embodiment 2
[0076] Example 2: PCR detection method for feed containing pigeon-derived components.
[0077] (1) DNA extraction:
[0078] Using phenol-chloroform extraction method: (refer to "Molecular Cloning Test Guide (Second Edition). Beijing: Science Press, 1995: 34-60")
[0079] ①Weigh 0.03g of the crushed sample of the substance to be detected into a 1.5ml centrifuge tube after autoclaving, and add 500μl of cell lysate to digest;
[0080] ② Add proteinase K (20mg / ml) 10μl, stir slowly to dissolve and mix well, if necessary, add proteinase K again, put it in a water bath at 50°C for digestion overnight, and mix well several times in due course;
[0081] ③ Cool the cell lysate to room temperature, add an equal volume of Tris-saturated phenol, and gently rotate the centrifuge tube to mix the two phases, so that the aqueous phase and phenol form an emulsion;
[0082] ④Centrifuge at 13000rpm / min at 4°C for 10min, carefully take out the centrifuge tube from the centrifuge, do not shake, ...
Embodiment 3
[0110] Preparation of Genomic DNA Extraction Reagent:
[0111] (1) Preparation of cell lysate:
[0112] ① 1M Tris-HCl 250mL:
[0113] Weigh 30.285g Tris, add water to make up to 250mL, add HCl to adjust the pH to 8.0, autoclave, and store at 4°C.
[0114] ②0.5mol / L EDTA:
[0115] Add 37.22g EDTA-Na2.2H2O to 100mL double-distilled water, stir vigorously, adjust the pH to 8.0 with NaOH (about 4gNaOH is needed), set the volume to 200mL, autoclave after aliquoting, and store at 4°C.
[0116] ③10%(m / v)SDS:
[0117] Dissolve 10g of SDS in 90mL of double-distilled water, heat to 68°C (to help dissolve a few drops of concentrated HCl, pH=7.2), add water to make up to 100mL, aliquot and store at room temperature without sterilization.
[0118] Take 1mol / L Tris-HCl (pH8.0) 2mL, 0.5mol / L EDTA (pH8.0) 40mL, 10% (m / v) SDS 10mL, add sterilized double-distilled water to 200mL .
[0119] (2) Preparation of 20 mg / ml proteinase K: 100 mg proteinase K was dissolved in 5 ml sterilized doubl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com