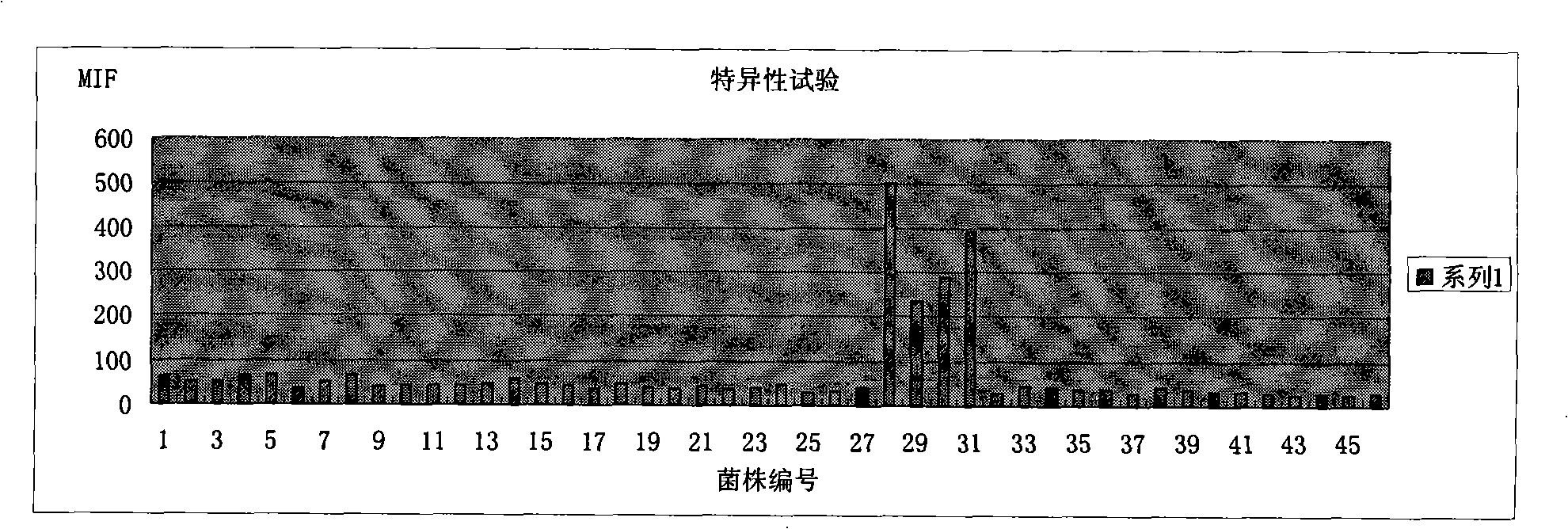
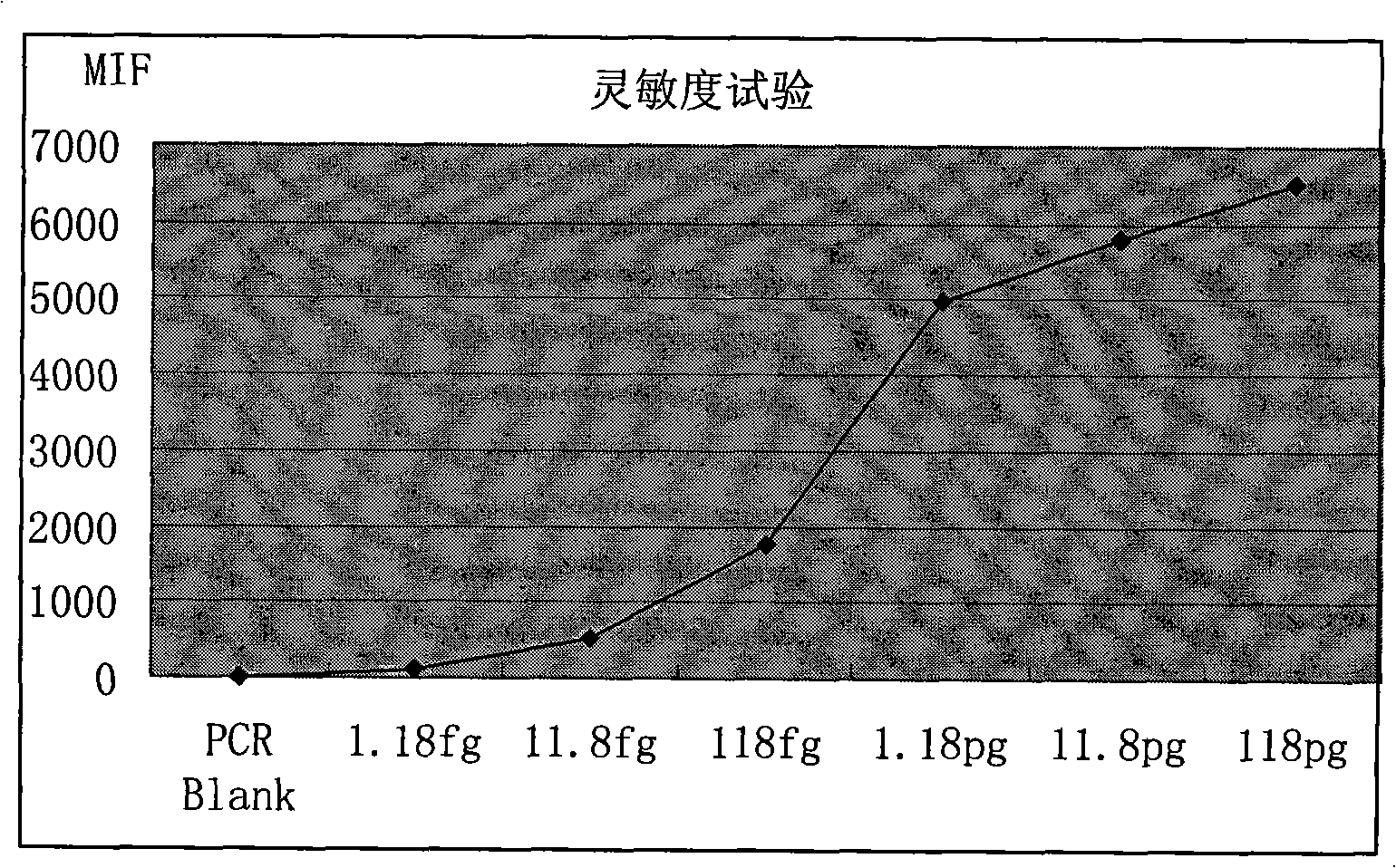
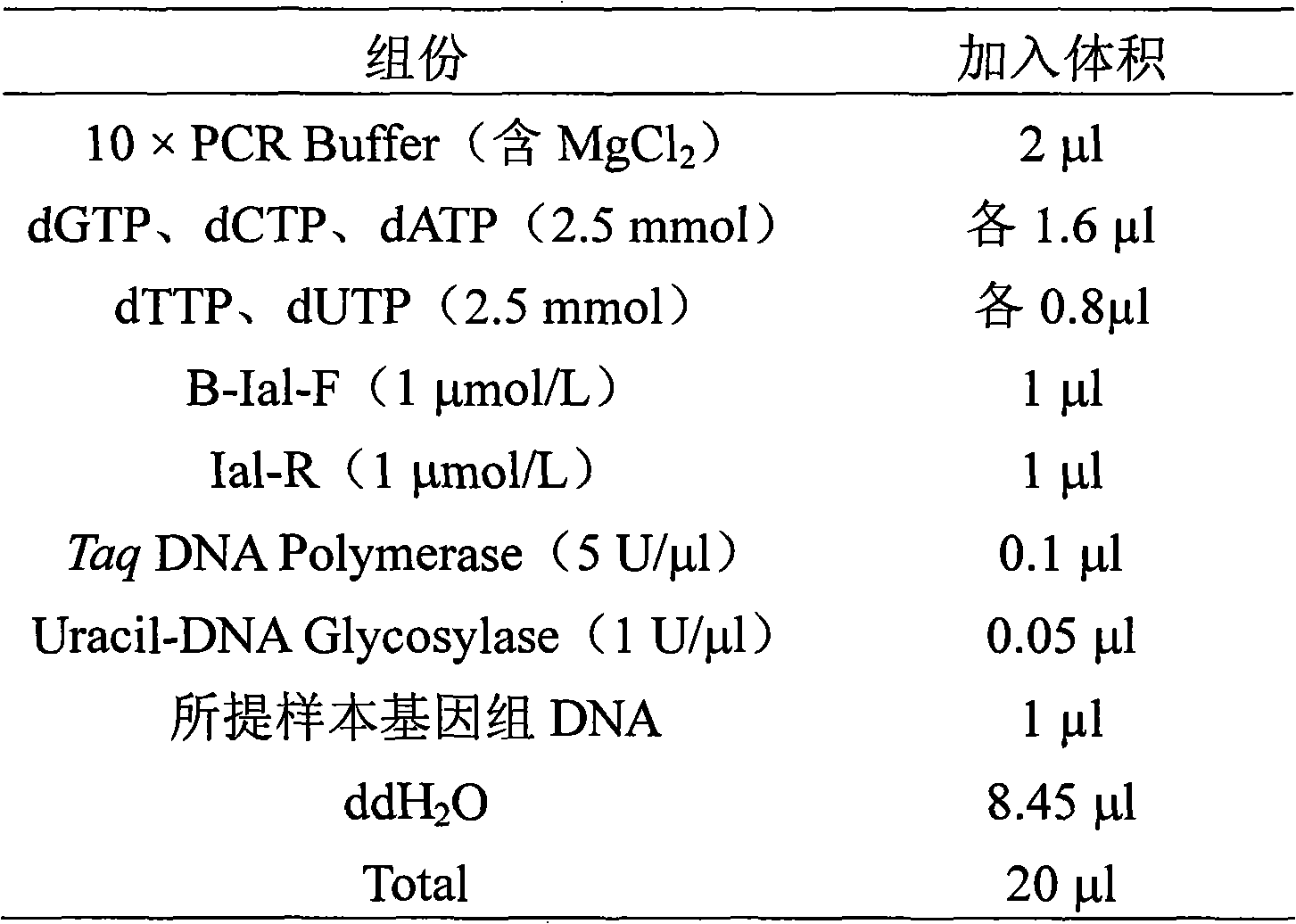
Method for detecting pathogenic shigella by using suspending chip technique
A Shigella, suspension chip technology, applied in biochemical equipment and methods, microbial determination/inspection, fluorescence/phosphorescence, etc., can solve problems such as inability to identify pathogenicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Design and synthesis of primers
[0035] 1) Sequence acquisition: Through genome-wide analysis of pathogenic Shigella, its specific gene Ial was selected as the target sequence, and the gene sequence was obtained from the GenBank public database, numbered AY206439;
[0036] 2) Design primers: use Primer Premier 5.0 software to design primers, the relevant parameters are: Tm value 55.0°C-59.0°C, GC value 40.0%-60.0%, PCR product size 100bp-500bp, primer size 22±3bp;
[0037] 3) Primer selection: Properly adjust the primers output by the software manually, increase or decrease a few bases, then perform online Blast comparison in GenBank, select primers with high specificity, and perform one of the 5′-end Biotin mark. The upstream primer sequence is B-Ial-F: 5′-Biotin-CTGGATGGTATGGTGAGGTTT-3′, the downstream primer is IpaH-R: 5′-AGGAGGCCAACAATTATTTCC-3′, and the amplified fragment size is 320bp;
[0038] 4) Primer synthesis: Shanghai Sangong synthesized the down...
Embodiment 2
[0039] Example 2: Design and synthesis of probes
[0040] 1) Sequence acquisition: designing probes in the non-primer region of the amplified fragment;
[0041] 2) design probes: adopt Primer Premier 5.0 software to design probes, select the HybridizationProbes command, design probes on the Anti-sense chain, and the parameters are the same as in Example 1;
[0042] 3) Probe selection: Properly adjust the probes output by the software manually, increase or decrease a few bases, then perform online Blast comparison in GenBank, select probes with high specificity, and perform NH at the 5′ end 2 -(CH 2 ) 12 Modification, the selected probe sequence is Ial-Probe: 5′-NH 2 -(CH 2 ) 12 -AATGTCCATCAAAACCCCACTC-3';
[0043] 4) Probe synthesis: Dalian TakaRa synthesized the above probes.
Embodiment 3
[0044] The preparation method of embodiment three suspension chips
[0045] 1. Materials:
[0046] 2. Method results:
[0047] 1. Dilute the amino-modified probe to 1mM (1nanomole / μl) with pure water;
[0048] 2. Shake the stored microspheres for 20 sec;
[0049] 3. Transfer 200μl microspheres to a brown EP tube;
[0050] 4. Collect the microspheres, centrifuge at 8000g for 1-2min;
[0051] 5. Discard the supernatant, add 50μl 0.1M MES (2-[N-Morpholino]ethanesulfonic acid, 2-(N-Morpholino)ethanesulfonic acid) solution pH 4.5, shake for 20sec;
[0052] 6. Add 2μl 1mM probe to the suspended microspheres, shake for 20sec;
[0053] 7. Prepare fresh 10mg / ml EDC (1-ethyl-3-[3dimethylaminopropyl]carbodiimide hydrochloride, 1-ethyl-3-3-dimethylaminopropyl carbodiimide), with dH 2 O;
[0054] 8. Add 2.5 μl of newly prepared 10 mg / ml EDC to the microsphere solution and shake;
[0055] 9. Incubate for 30 minutes at room temperature and in the dark;
[0056] 10. Prepare fresh 10m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com