Method for detecting Shigella by using suspension chip technology
A Shigella, suspension chip technology, applied in biochemical equipment and methods, microbial determination/inspection, fluorescence/phosphorescence, etc., can solve the problem of difficult to control false positive and false negative rates, immature technology, cross-contamination, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Design and synthesis of primers
[0053] 1) Sequence acquisition: Through genome-wide analysis of Shigella, the specific gene IpaH was selected as the target sequence, and the gene sequence was obtained from the GenBank public database, numbered SHFIPAHX;
[0054] 2) Design primers: use Primer Premier 5.0 software to design primers. The relevant parameters are: Tm value 55.0°C-59.0°C, GC value 40.0%-60.0%, PCR product size 100bp-500bp, primer size 22±3bp;
[0055] 3) Primer selection: Properly adjust the primers output by the software manually, increase or decrease a few bases, then perform online Blast comparison in GenBank, select primers with high specificity, and perform one of the 5′-end Biotin mark. The upstream primer sequence is B-IpaH-F: 5′-Biotin-CTTGACCGCCTTTCCGATAC-3′, the downstream primer is IpaH-R: 5′-ACTCCCGACACGCCATAGA-3′, and the amplified fragment size is 421bp;
[0056] 4) Primer synthesis: Shanghai Sangong synthesized downstream primer ...
Embodiment 2
[0057] Example 2: Design and synthesis of probes
[0058] 1) Sequence acquisition: designing probes in the non-primer region of the amplified fragment;
[0059] 2) design probes: adopt Primer Premier 5.0 software to design probes, select the HybridizationProbes command, design probes on the Anti-sense chain, and the parameters are the same as in Example 1;
[0060] 3) Probe selection: Properly adjust the probes output by the software manually, increase or decrease a few bases, then perform online Blast comparison in GenBank, select probes with high specificity, and perform NH at the 5′ end 2 -(CH 2 ) 12 Modification, the selected probe sequence is Ipah-Probe: 5′-NH 2 -(CH 2 ) 12 -CGGAGATTGTTCCATGTGAGC-3';
[0061] 4) Probe synthesis: Dalian TakaRa synthesized the above probes.
Embodiment 3
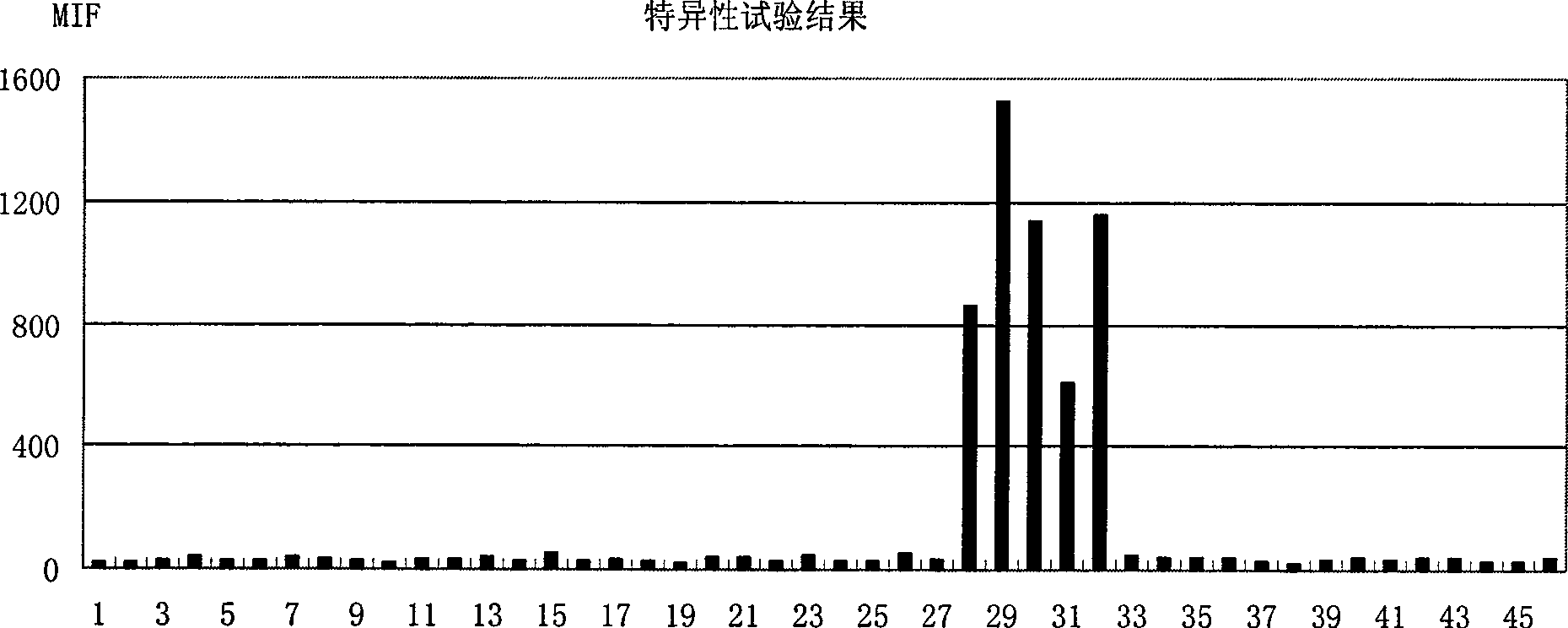
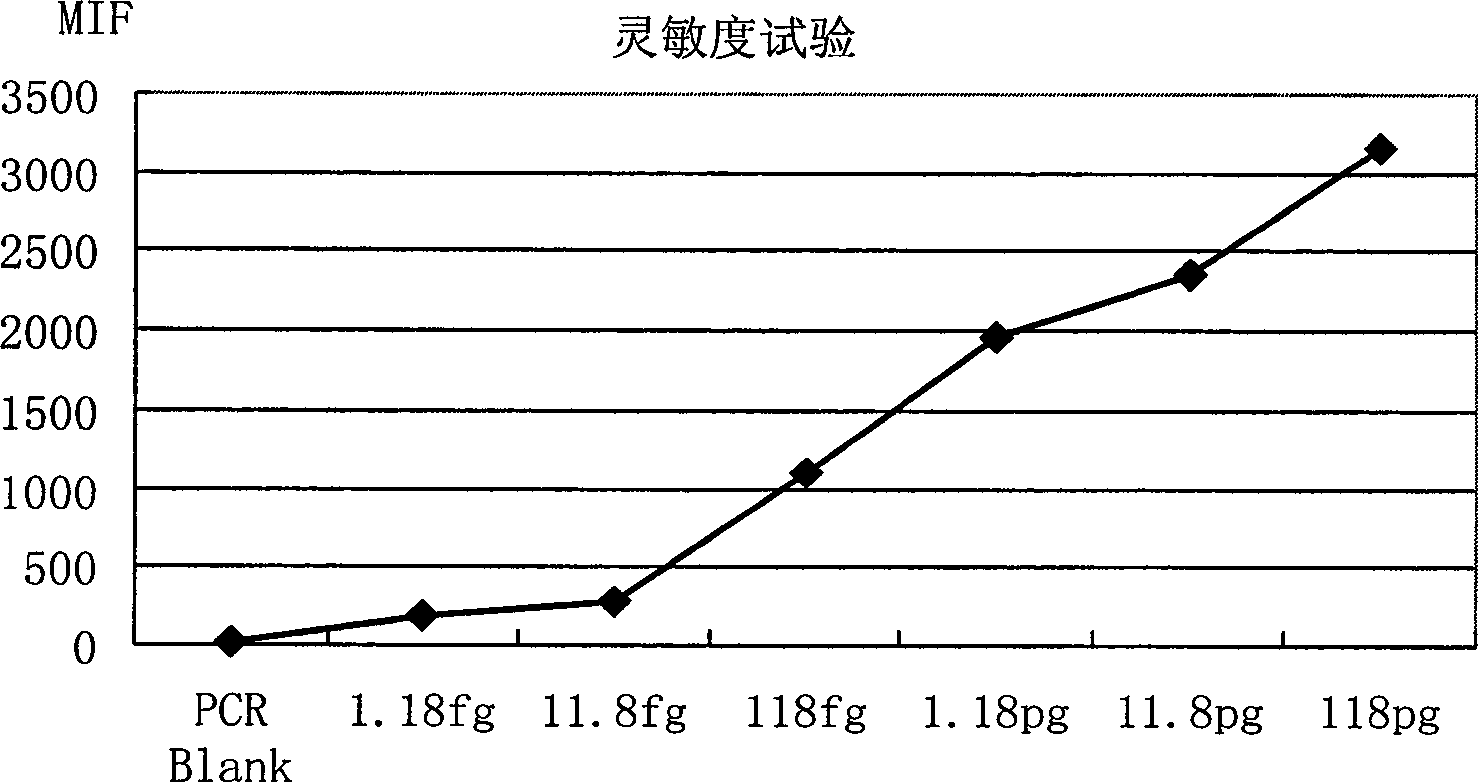
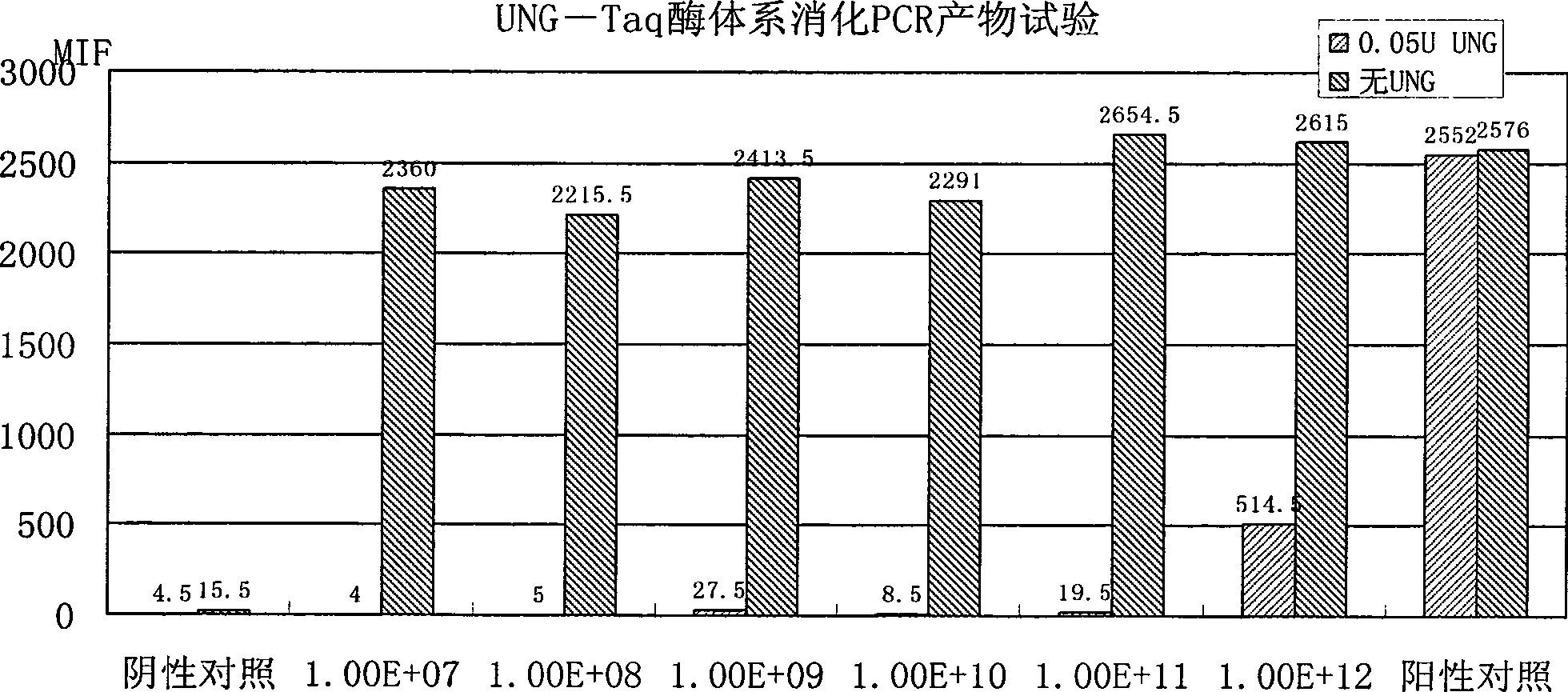
[0062] Example 3: Suspension Chip Rapid Detection Method for Shigella
[0063] 1) TIANamp Bacteria DNA kit was used to extract the genomic DNA of the sample to be tested.
[0064] 2) PCR amplification of the target fragment, the reaction conditions are as follows:
[0065] Table 2 PCR reaction system
[0066]
[0067] The PCR reaction program was: denaturation at 94°C for 3 min; 35 cycles of 30 sec at 94°C, 30 sec at 53°C, 40 sec at 72°C; finally, extension at 72°C for 3 min.
[0068] 3) Hybridization: Hybridization solution composition: 33.3 μl 1.5×TMAC (tetramethylammonium chloride, tetramethylammonium chloride), 5 μl of the above PCR product, 5000 probe-coupled microspheres were added, and 1×TE to make up the volume to 50 μl. The hybridization solution was denatured at 95°C for 4 minutes, and hybridized at 52°C for more than 15 minutes.
[0069] 4) Detection: Transfer the above-mentioned hybridized solution to a 96-well filter plate, collect the microspheres by vacuum...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com