Molecular genetic identification technique of Eriocheir sinensis and Eriocheir japonica
A technology of Chinese mitten crab and mitten crab, which is applied in the fields of biodiversity conservation and genetics, and can solve the problems of lack of molecular marker technology and methods for identifying mitten crabs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
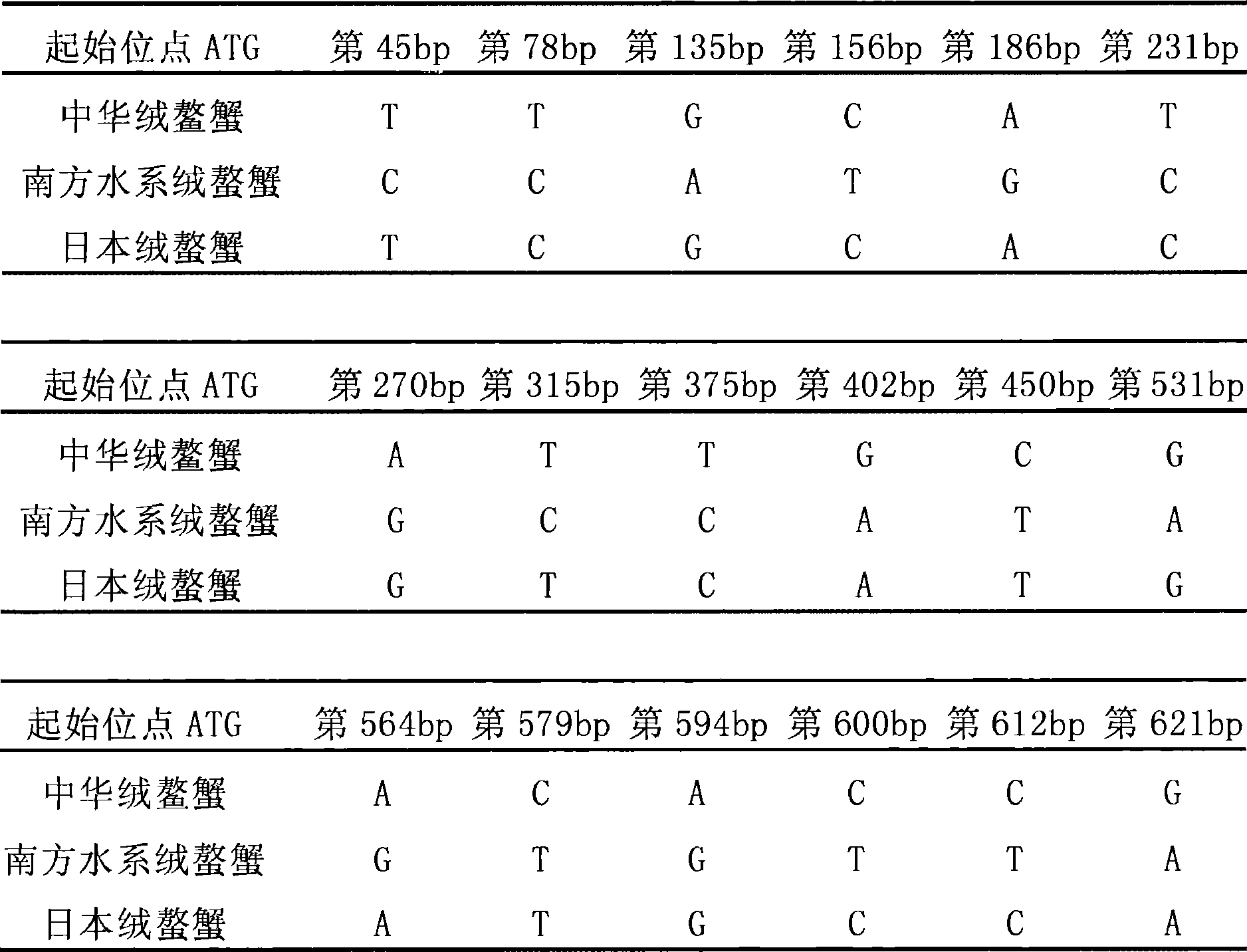
Image
Examples
Embodiment 1
[0107] Example 1. Sample Selection
[0108] A total of 125 samples of Chinese mitten crabs from different water systems, 40 samples of mitten crabs from southern Chinese water systems (Nanliu River in Guangxi and Pearl River in Guangdong), and 15 samples of Japanese mitten crabs from Hokkaido, Japan were selected for subsequent experiments . The inventor has selected a sufficient number of mitten crabs, so the obtained results are statistically significant, accurate and reliable.
Embodiment 2
[0109] Example 2.DNA Extraction
[0110] Take 0.2g of the long segment muscle of the third step of each crab, put it into a 1.5ml centrifuge tube, add 400μL STE buffer solution (30mmol / L Tris-HCl, pH8.0, 200mmol / L EDTA, 50mmol / L NaCl) . After mixing, add 90 μL of SDS with a concentration of 10% and 10 μL of proteinase K at 20 mg / mL, digest at 55°C for 8-10 hours; add an equal volume of saturated phenol, rotate slowly on the rotor for 1 hour, centrifuge at 10,000 r / min for 8 minutes, and absorb the supernatant Add an equal volume of mixed solution (phenol: chloroform: isoamyl alcohol = 25:24:1); rotate slowly for 30 min, centrifuge at 10,000 r / min for 8 min, take the supernatant and add an equal volume of chloroform and isoamyl alcohol mixed solution (chloroform : isoamyl alcohol=24:1), rotate slowly for 5min, centrifuge at 10,000r / min for 5min, take the supernatant, add 2 times the volume of absolute ethanol to precipitate DNA, then wash with 70% ethanol, dry, add 300μL Diss...
Embodiment 3
[0111] Example 3.PCR amplification and product cloning, sequencing
[0112] According to the Chinese mitten crab mitochondrial COII gene sequence reported in the 2001 Journal of Nanjing Normal University Natural Science Edition, Volume 24, Phase 4, Page 485-490, a pair of degenerate primers designed to amplify the COII gene are:
[0113] COII-F: 5'-CATCACCTTGTCAAGGTGAAA-3' (SEQ ID NO: 4);
[0114] COII-R: 5'-CATGGTCAGTCTCAGGATTCA-3' (SEQ ID NO: 5).
[0115] The crab genomic DNA prepared in Example 1 was used as a template, and the above-mentioned COII-F and COII-R were used as primers for PCR amplification. The PCR reaction volume is 50 μL, containing 5 μL PCR buffer (10 mmol / L Tris-HCL, pH9.0, 50 mmol / L KCL, 30 mmol / L MgCL2, 0.01% gelatin), 2 μL dNTP mixture (each dNTP 0.1 mmol / L ), 4 μL of primers (0.2 μmol / L), 2 μL of genomic DNA (50 ng / μl), 2 μL of Taq enzyme (2 IU), and 35 μL of distilled water.
[0116] The amplification reaction conditions were: pre-denaturation at 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com