Candida utilis expression vector and construction method thereof
A technology for producing Candida utilis and an expression vector, which is applied in the fields of expressing exogenous proteins and molecular biology to save human, material, and financial resources.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1: Primer design
[0019] According to the gene sequence of C.utilis published in GenBank combined with pBR322 plasmid sequence characteristics, primers were designed and suitable restriction sites were introduced. Among them, primers P5 and R5 were designed according to literature. The primers are as follows:
[0020] P1: 5’-GGATATCTTACAGCGAGCACTCAA-3’ (Introduction of Eco RV restriction site)
[0021] R1: 5'-AGGTACCGCTAGCTCTAGAATGTTGTTTGT-3' (introduction of Kpn I, Nhe I and Xba I restriction sites)
[0022] P2: 5’-ttctagagctagcggtacctatgacttttat-3’ (introduction of Xba I, Nhe I and Kpn I restriction sites)
[0023] R2: 5'-gggatccACGTGTAATACCAGG-3' (introduction of Bam HI restriction site)
[0024] P3: 5’-cgatatcTGCCAGTAGTCATATGC-3’
[0025] R3: 5’-cgatatcTGACTTGCGCTTACTAG-3’ (Introduce Eco RV restriction sites at the 5’ ends of both primers)
[0026] P4: 5'-CgtcgacAGTAAGTATGAAAAGAGC-3' (Introduction of Sal I site)
[0027] R4: 5’-GggatccGG GTTTGGTCTATGTTGCT-3’ (i...
Embodiment 2
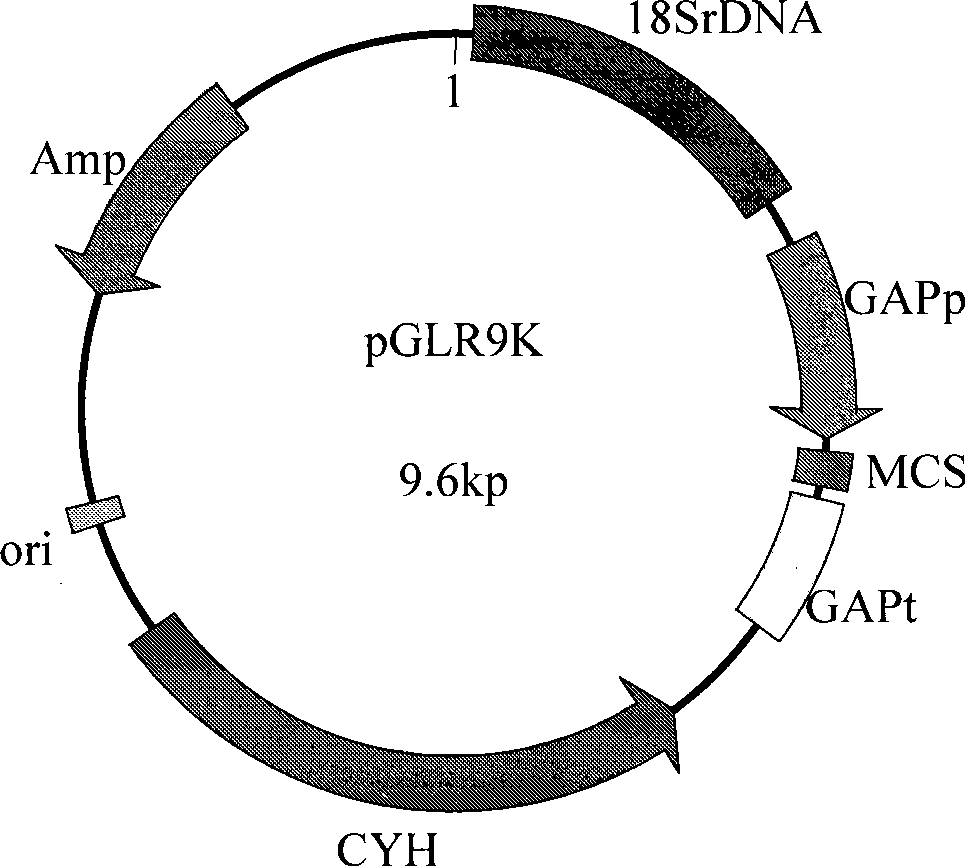
[0032] Example 2: Construction of homologous integration expression vector pGLR9K
[0033] The C.utilis genome was used as a template and P4 / R5 was used as primers to perform PCR to obtain upstream fragments of the L41 gene. Similarly, the C.utilis genome was used as a template and P5 / R4 as primers to perform PCR to obtain downstream fragments of the L41 gene. The two fragments were recovered separately, and the two fragments were mixed in equal amounts as templates, and PCR was performed with P4 / R4 as primers to obtain the CYH resistance gene. The PCR reaction conditions were: 94°C for 5 min; 94°C denaturation for 30 seconds, 56°C annealing for 30 seconds, 72°C extension for 2 minutes, 30 cycles, 72°C extension for 10 minutes. The recovered target fragment was connected to the vector pMD19-T simplevector (named pT-CYH) for sequencing analysis.
[0034] Use C.utilis genome as template and P1 / R1 as primers to perform PCR to obtain yeast GAP promoter (GAP-p) gene fragment; also use ...
Embodiment 3
[0039] Example 3: Construction of recombinant expression vector containing target gene
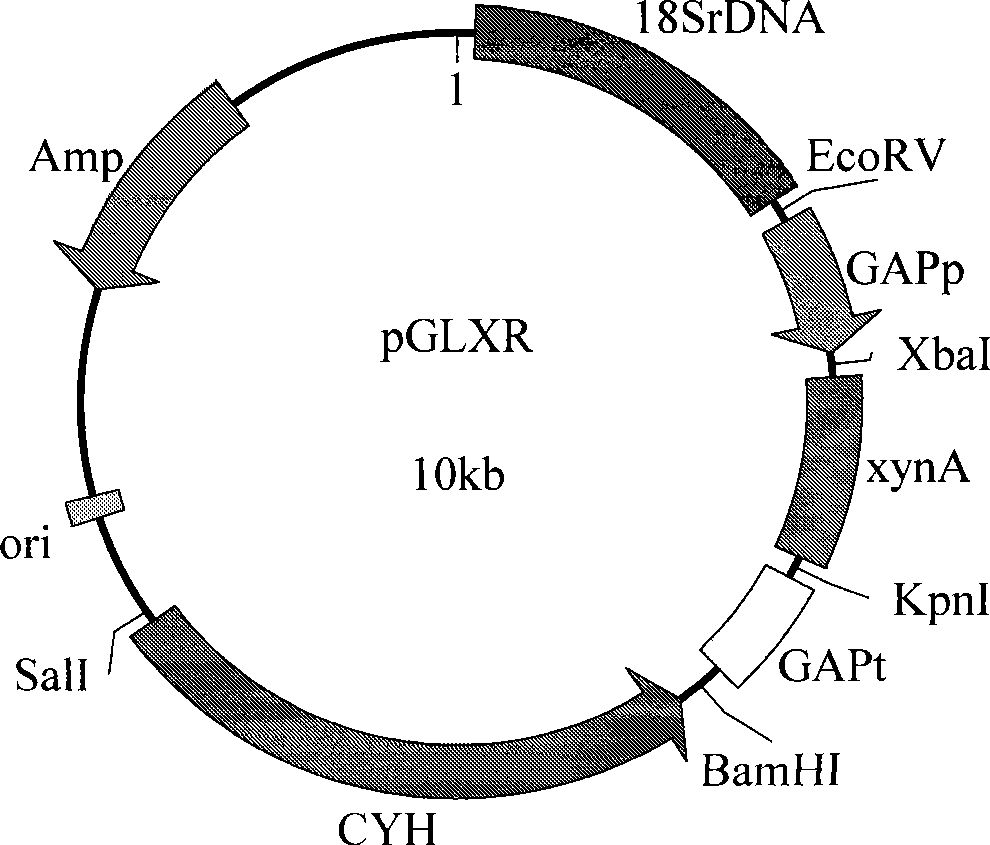
[0040] Using plasmid pUC19-XA as a template and P6 / R6 as primers, PCR was performed to obtain the xynA target gene fragment. The PCR conditions were: 94°C for 5 min; 94°C denaturation for 30 seconds, 58°C annealing for 30 seconds, 72°C extension for 1.5 minutes, 30 cycles; 72°C extension for 10 minutes. The target gene was recovered and cloned into the vector pMD19-T simple vector (named pT-XA) for sequencing analysis.
[0041] The pT-XA plasmid was extracted, digested with Xba I / Kpn I, and the target fragment was recovered and constructed into the vector pGLR9K multiple cloning site to obtain the plasmid pGLXR. Transform E. coli DH5α, screen and identify positive clones and save them for later use. See attached figure 2
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com