Novel pectate lyase and method of use for bio-scouring
A technology of Bacillus subtilis and pectic acid, applied in textiles and papermaking, biochemical treatment of enzymes/microbes, lyase, etc., can solve the problem of not being able to remove fiber-pigment groups
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0401] Example 1. Synthesis and purification of enzyme
[0402] Using the aprE promoter for efficiently transcribing genes and the AprE signal sequence that enables the efficient secretion of pectate lyase into the medium, pectate lyase derived from a derivative of Bacillus subtilis subtilis strain 168 is produced in Bacillus subtilis. Using forward oligonucleotide primer 5'-TTCAGCGAAGACAGCGCGCAGGCAGCTGATTTAGGCCACCAGACGTTGGG-3' (SEQ ID NO: 3) and reverse primer 5'-TTTTCAAAGCTTTAATTTAATTTACCCGCACCCGCTTGATTT-3' (SEQ ID NO: 4), using PCR from chromosomal DNA (using standard phenol The extraction technique was isolated from the derivative of Bacillus subtilis subtilis strain 168) to amplify the pel gene. The forward primer fused the coding sequence of the AprE signal sequence to the coding sequence of the mature pel gene in frame. The forward primer also includes a BbsI restriction site, which generates an overhang sequence compatible with the BssHII site after cleavage. The reverse p...
Embodiment 2
[0425] Example 2. Characterization of pectate lyase
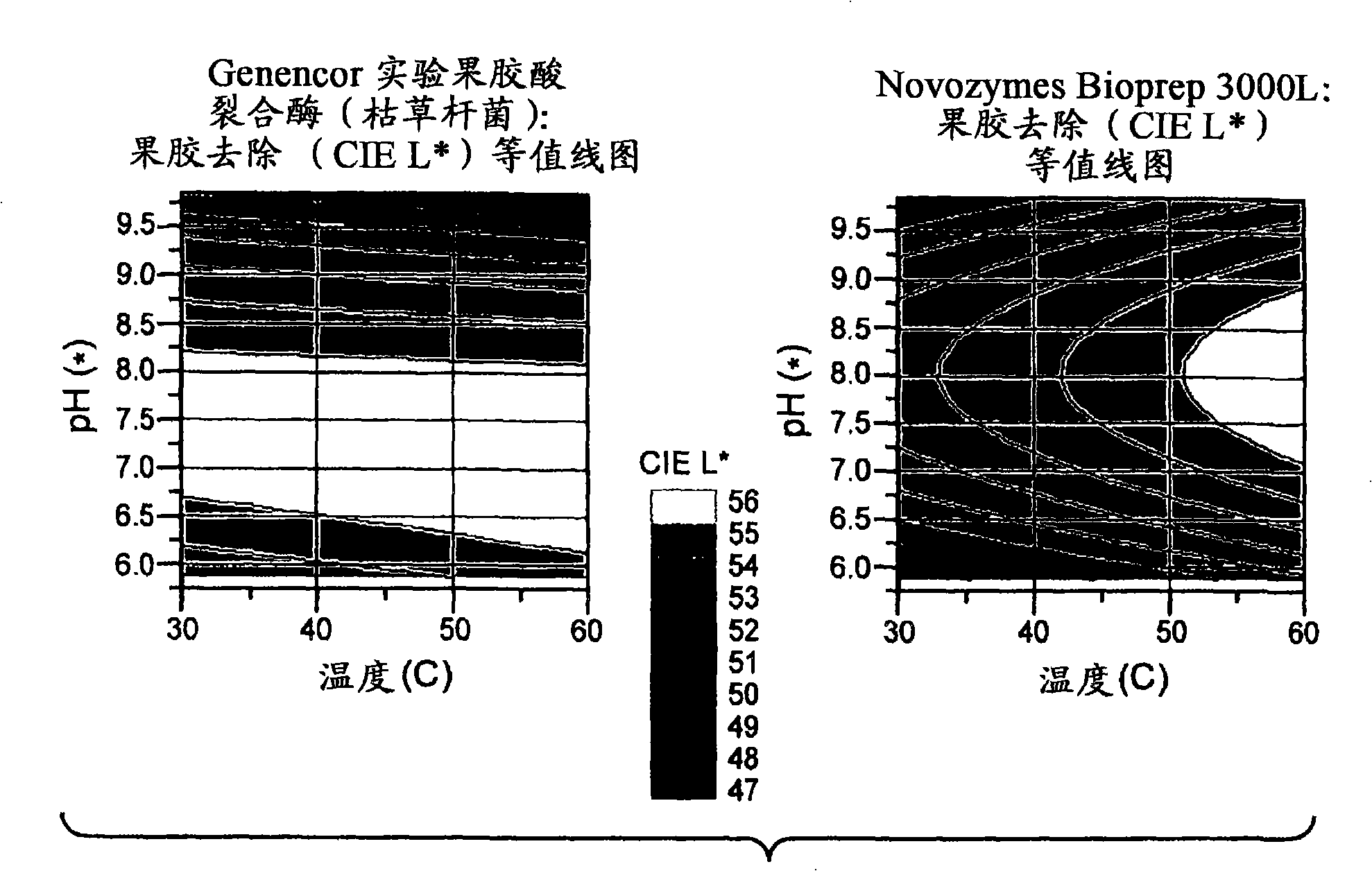
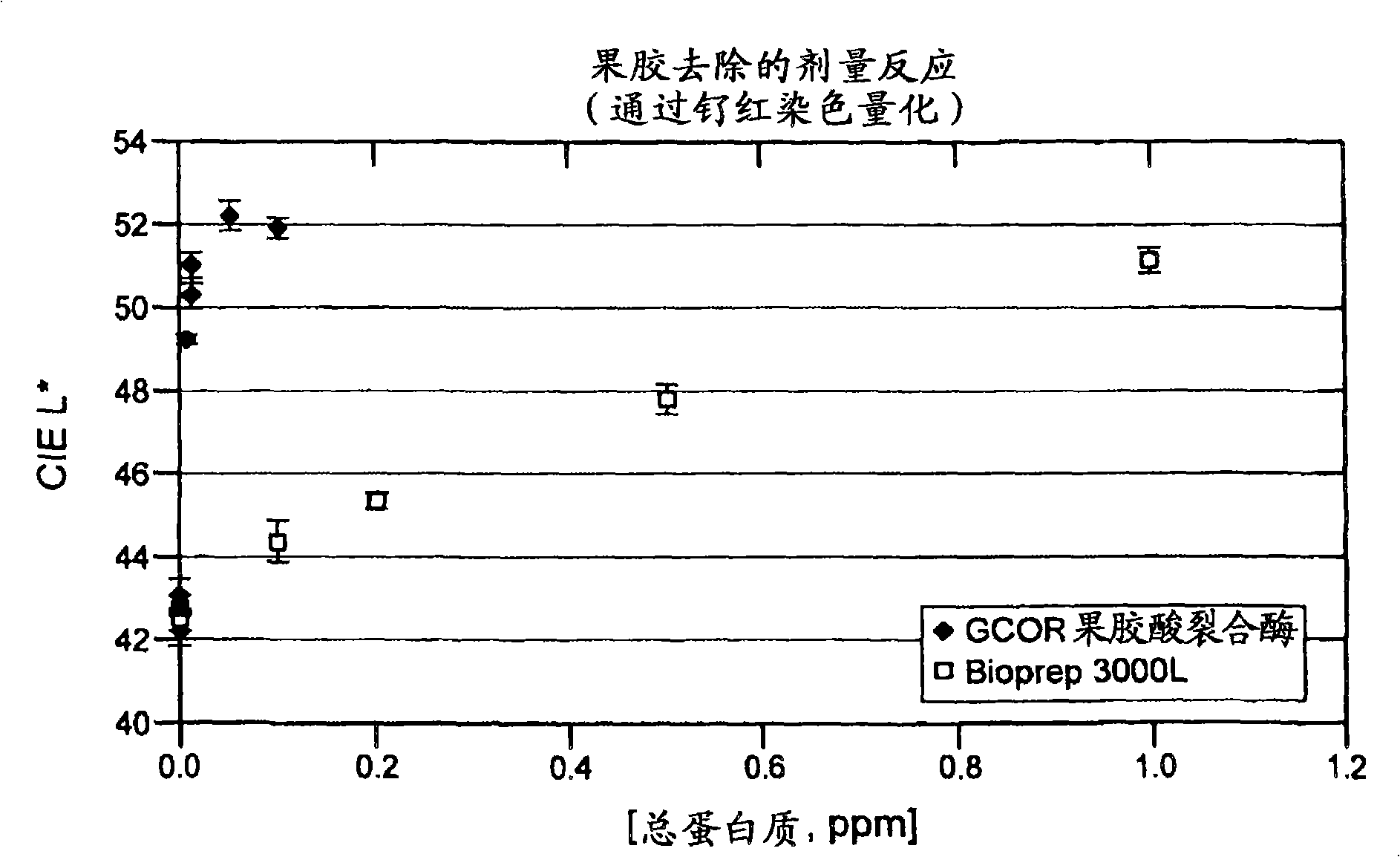
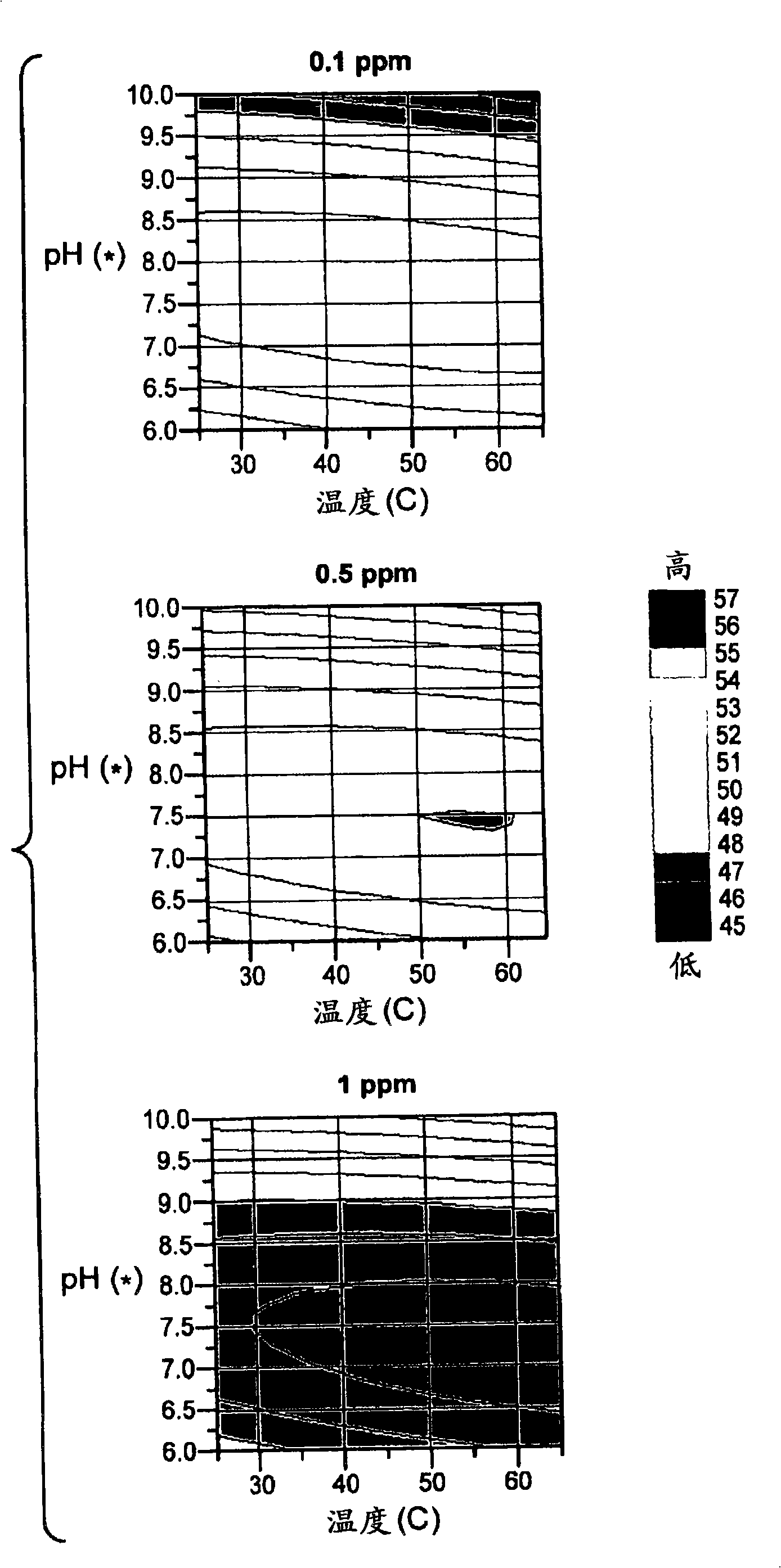
[0426] Under the following conditions, the pectin removal performance of Bacillus subtilis pectate lyase (SEQ ID NO: 1) was compared with that of a commercially available pectate lyase, Bioprep 3000L (Novozymes).
[0427] Pectate lyase
Total protein * mg / mL
TCA protein * (mg / mL)
Exemplary Bacillus subtilis of Example 1
87.8
53.4
Bioprep 3000L
(Novozymes)
44.0
35.5
[0428] " * "This result is calculated based on 6.35g protein / g nitrogen. In addition, this result is an estimate of total protein.
[0429] Material: The dirt carrier used was a desizing but unbleached fabric from Testfabrics ArmyCarded Cotton Sateen (Style#428). The Bacillus subtilis pectate lyase and Novozymes Bioprep 3000L pectate lyase of Example 1 were utilized at 2 ppm total protein. Analysis of various pH 50mM BIS-TRIS propane (BTP) buffers. Use HCl to adjust the buffe...
Embodiment 3
[0441] Using acetic acid, the pectate lyase of Example 1 was formulated with propylene glycol (40%), Proxel (0.25%) (1,2-benzisothiazolin-3-one), and pH 6.25±0.1. The test was performed by incubating at 37°C for 20 minutes. Use CaCl 2 Glycine buffer with polygalacturonic acid substrate for this endpoint measurement.
[0442]
Fermented pectin
Acid lyase
Test
Volume
(μl)
pH 10
235nm
OD
Blind 210
pH 10
235nm end
OD
pH 10
APSU
Sample
APSU / ml
20040772UFC
500
0.712
0.124
0.588
0.137458362
91638.91
20040773UFC
500
0.613
0.124
0.489
0.116094893
77396.6
End of 20040772
500
0.336
0.124
0.212
0.056320337
37546.89
End of 20040773
500
0.467
0.124
0.343
0.08458917
56392.78
20040396
Whole broth
500
0.415
0.124
0...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com