Transgene report pig detecting Cre site special recombinase active and culturing method
A culture method and recombinase technology, applied in the field of zoology, can solve problems such as large differences in physiological characteristics and inability to replicate human diseases well, and achieve the effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
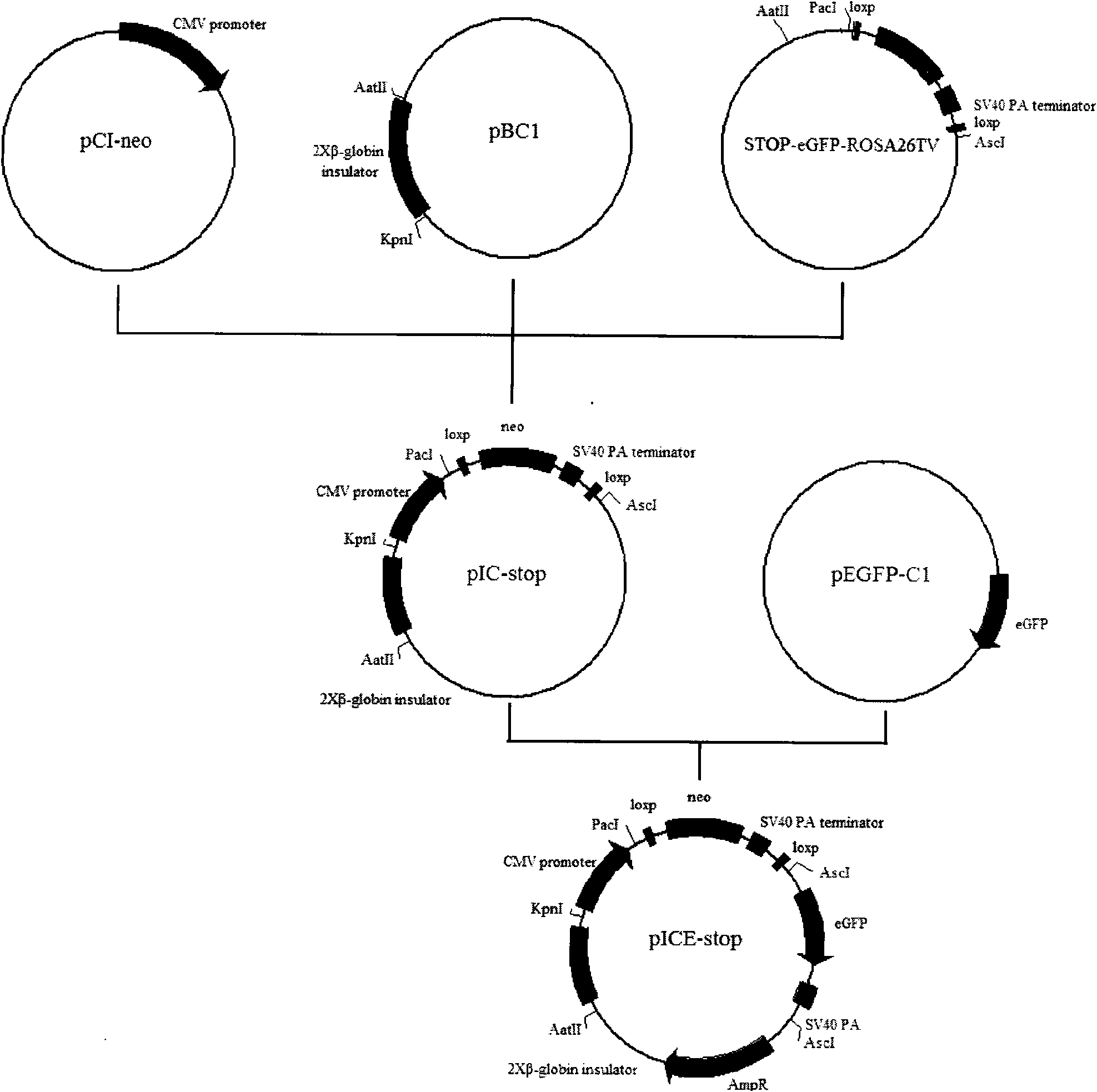
[0058] Insulator, CMV promoter, amplification of green fluorescent protein gene
[0059] Using PCI-Neo as a template, add primers C1 and C2 to amplify the CMV promoter through PCR cycle, the full-length sequence is 1022bp
[0060] Using PBC-1 as a template, add primers I1 and I2 to amplify the insulator through PCR cycle, the full length is 2450bp
[0061] Using pEGFP-C1 as a template, adding primers E1 and E2 to amplify the eGFP gene through PCR cycles, the full-length sequence is 804bp.
[0062] The amplified sequence was connected to PMD-18T-Simple (takara company), and entrusted to Beijing Tiangen Company to perform sequencing. After digesting the sequenced correct fragments, they were respectively connected into the STOP-eGFP-ROSA26TV vector and identified by enzyme digestion After the sequencing was correct, it was named pICE-STOP. After expanding the culture, the plasmid was extracted using Tiangen’s large-scale plasmid extraction kit, and linearized with NheI single-e...
Embodiment 2
[0064] Cell transfection and selection
[0065] Transfection of embryonic fibroblasts using liposome-mediated DNA, steps. Follow the instructions of lipofectin2000 (Invitrogen). The day before the transfection, the primary cultured embryonic fibroblasts were resuscitated into a 60mm plate with antibiotic-free culture medium, and the cells could be transfected when the cells reached 70-80% confluency. 24 hours after transfection, cells were passaged by trypsinization 1:21. 48 hours after transfection, G418 selection medium (concentration: 300 μg / ml) was added. When a single colony of cells appears, remove the culture medium, wash once with PBS at 39°C, put a self-made glass cell cloning ring on the cell colony, add an appropriate amount of trypsin to the cloning ring, and digest for about 1 minute , adding selection medium to terminate the reaction. Carefully blow the cells with a pipette and transfer to a 24-well plate for expanded culture. After the cells are overgrown, t...
Embodiment 3
[0067] Identification at the Cellular Level -
[0068] 1. PCR detection
[0069] The obtained neo gene-positive cell genome was used as a template, and primers C1 and E2 were used to amplify, and the amplified sequence included CMV promoter, Loxp-neo-stop-Loxp sequence, and EGFP gene. It proves that the target gene is fully integrated into the genome of porcine embryonic fibroblasts.
[0070] 2. Fluorescence observation
[0071] In order to verify whether the constructed reporter cell line conditionally expressing green fluorescent protein can effectively monitor the activity of Cre recombinase, the screened cell clones were expanded and cultured, and then infected with adenovirus expressing Cre recombinase to verify the activity of Cre recombinase. Validation of construction of reporter cell lines.
[0072] 3. RT-PCR
[0073] Three days after the obtained cell clones were infected with the adenovirus, the total RNA of the cells was extracted for RT-PCR reaction to further...
PUM
| Property | Measurement | Unit |
|---|---|---|
| weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com