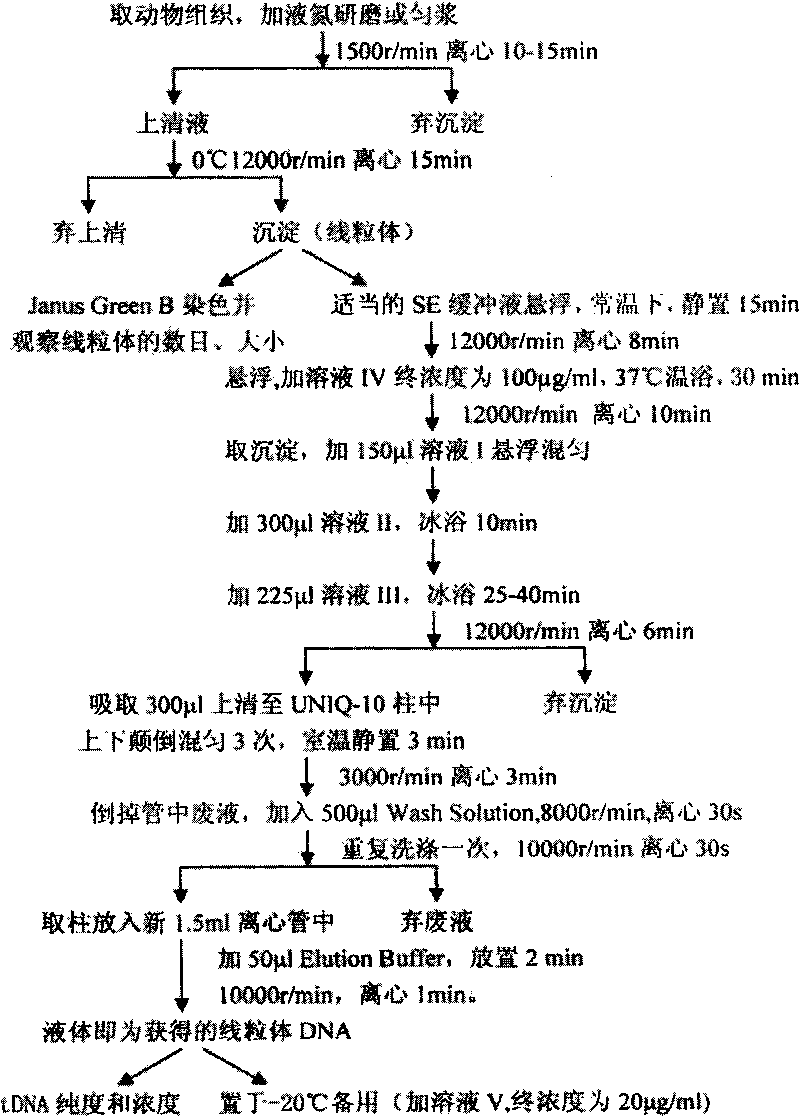
Method for extracting and purifying columnar high-purity animal mtDNA
A purification method and high-purity technology, which is applied in the field of nucleic acid extraction and purification in molecular biology, can solve the problems of harmfulness and residues in environmental operators, and achieve high-purity results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Extraction of frog mitochondrial DNA
[0024] (1) Take 2-15g of liver, gonad or muscle of black-spotted frog, chop it into a small amount of SE homogenization buffer, add about 20-30mL of SE solution, homogenize up and down with an electric homogenizer at 1500r / min for 30 seconds, and filter , pipette the homogenate into a centrifuge tube.
[0025] (2) Centrifuge at 1500r / min for 15min to absorb the supernatant. Centrifuge at 12000r / min for 20min to take the precipitate, add 10ml SE solution to suspend the sediment, repeat centrifugation once, add 10ml STM solution to suspend mitochondria, add solution IV to make the final concentration 100μg / ml, warm bath at 37℃, 30min, centrifuge at 12000r / min for 10min, discard the supernatant solution, add 4ml SE solution to suspend mitochondria, and then transfer to four 1.5ml Eppendorf tubes, 1ml in each tube.
[0026] (3) Centrifuge at 12000r / min for 8min to discard the supernatant, add 150μl solution I to each tube,...
Embodiment 2
[0031] Example 2: Extraction of fish mitochondrial DNA.
[0032] (1) Take 5-6g fish liver or gonad tissues, rinse them in a small amount of SE solution, cut them into pieces, add about 50ml SE, and homogenize them up and down with an electric homogenizer at 1500r / min for 30s.
[0033] (2) The homogenate was centrifuged at 1000r / min for 15min to absorb the supernatant, and then centrifuged at 12000r / min for 20min, and the precipitate was mitochondria. Add 4ml of STM solution to suspend the mitochondria, add solution IV to make the final concentration 100μg / ml, incubate at 37°C for 30min, centrifuge at 12000r / min for 10min, discard the supernatant; add 4ml of SE solution to suspend the mitochondria, transfer to four 1.5ml Eppendorf tubes.
[0034] (3) Centrifuge at 12000r / min for 10min, discard the supernatant, add 150μl of solution I to each tube, suspend the precipitate, add 300μl of newly prepared solution II, mix well, and ice-bath for 10min, then add 225μl of cold solution ...
Embodiment 3
[0038] Example 3: Extraction and purification of honeybee mitochondrial DNA
[0039] (1) Take fresh honey bee or alcohol soaked specimens, add liquid nitrogen to grind, add about 5-10mL SE solution, mix well, filter, and suck the homogenate into a centrifuge tube.
[0040] (2) Centrifuge at 1500r / min for 15min to absorb the supernatant. Centrifuge at 12000r / min for 20min to take the precipitate, add 2ml of STM solution to suspend, add solution IV to make the final concentration 100μg / ml, warm bath at 37°C for 30min, centrifuge at 12000r / min for 10min, discard the supernatant, add 2mL of SE solution to suspend, and then transfer to 2 1.5ml Eppendorf tubes, 1ml per tube.
[0041] (3) Centrifuge at 12000r / min for 8min to discard the supernatant, add 150μL of solution I to each tube, pipette the precipitate evenly, add 300μL of freshly prepared solution II, mix well, and ice-bath for 10min, add 225μL of cold solution III each, mix well, and ice-bath 60min, centrifuged at 12000r / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com