Overall length cDNA sequence of micro-capsule algae toxins degrading enzyme MlrA, coded amino acid and application
A microcystin and enzyme-degrading technology, which is applied in the fields of biotechnology applications and environmental protection, can solve the problems of high cost, inability to completely remove MC, and heavy workload, and achieve the goal of maintaining vitality, maintaining detoxification ability, and reducing damage Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
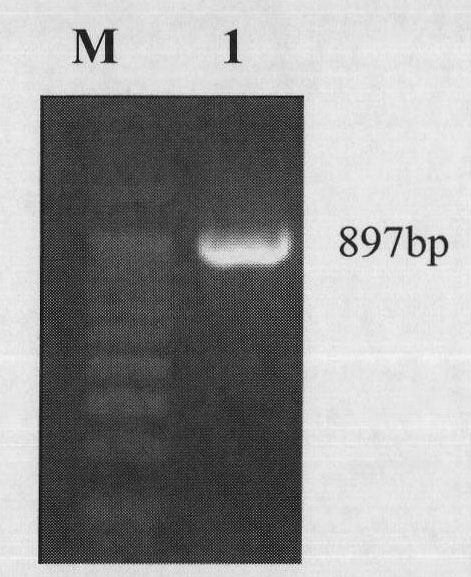
[0058] Example 1 Obtaining of cDNA sequence of MlrA gene and its encoded amino acid sequence
[0059] 1. Sample Collection and Processing
[0060] The water samples used in the experiment were collected from Xiangang Reservoir, Huizhou City, Guangdong Province, Minghu Lake, Jinan University, Guangzhou City, and Guangdong Tilapia Breeding Farm, and fish gut samples were collected from Xiangang Reservoir, Huizhou City, Guangdong Province.
[0061] Water samples were collected from surface water bodies at various locations with sampling bottles.
[0062] The fish intestines were killed on the spot, and the hindguts (about 10 cm) were taken, and the excreta in the intestines were picked out and mixed with 15 ml of distilled water to obtain fish intestine samples.
[0063] After mixing the above water sample and fish intestinal sample, filter through a medium-speed qualitative filter paper to remove impurities, centrifuge at 12000rpm for 1min, collect the microbial mixture, and st...
Embodiment 2
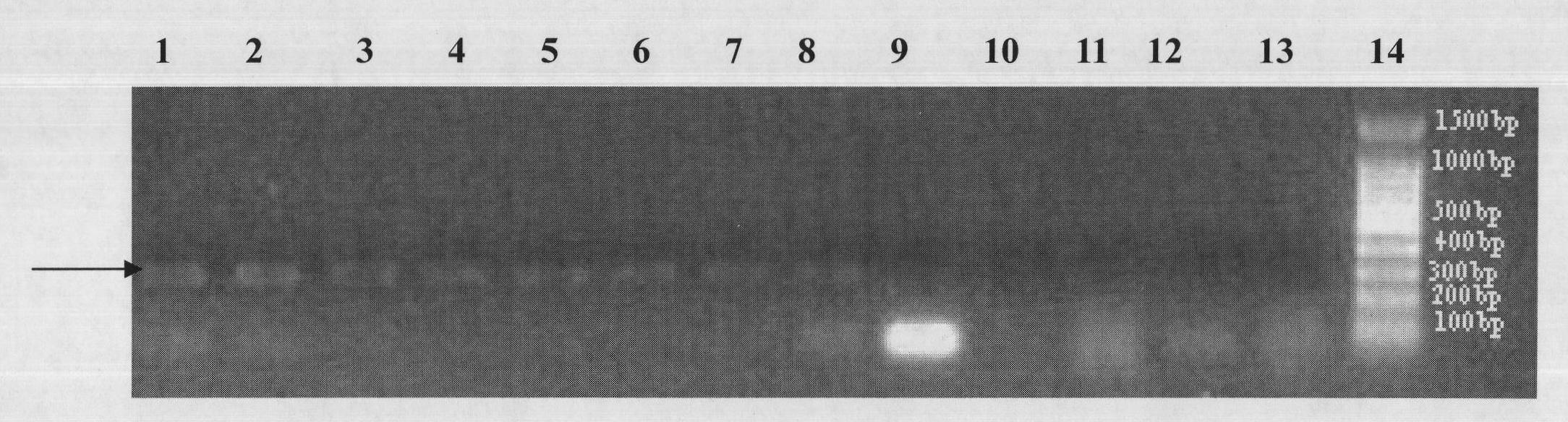
[0081] Example 2 MlrA detection results in water bodies and fish bodies in different seasons
[0082] 1. Sample Collection and Processing
[0083] The water samples used in the experiment were collected from Xiangang Reservoir, Huizhou City, Guangdong Province, Minghu Lake, Jinan University, Guangzhou City, and Guangdong Tilapia Breeding Farm, and fish gut samples were collected from Xiangang Reservoir, Huizhou City, Guangdong Province.
[0084] In March, April, May, July, August, October, November and December, water samples and fish intestinal samples were collected respectively.
[0085]Water samples were collected from surface water bodies at various locations with sampling bottles.
[0086] The fish gut samples were killed on the spot, and the hindguts (about 10 cm) were taken, and the excreta in the guts were picked out and mixed with 15 ml of distilled water to obtain the fish gut samples.
[0087] After the water samples and fish intestinal samples in the water envir...
Embodiment 3
[0105] Example 3 Preparation of MlrA crude enzyme preparation expressing MlrA protein
[0106] 1. Construction of recombinant expression vector pET3c-MlrA
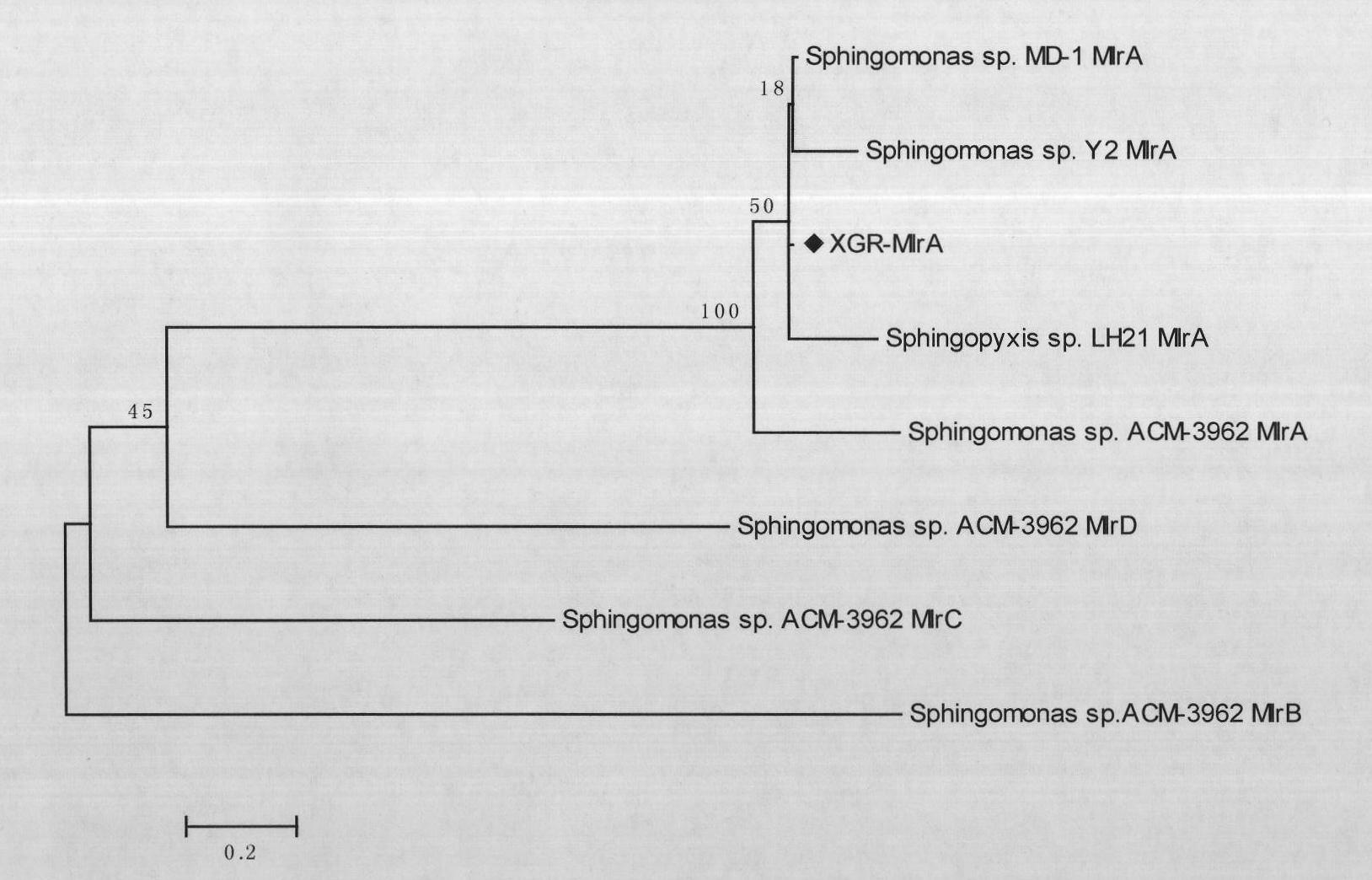
[0107] The full-length cDNA sequence of the Sphingomonas MlrA gene obtained in Example 1 was cloned, and the PCR product was purified using H.Q.&.Q.Gel Extraction Kit II.
[0108] Take 50 μL of the E.coli DM5α preservation solution containing the pET3c plasmid, inoculate it in 4 mL of LB liquid medium (containing Amp), and cultivate overnight at 37°C with shaking at 280 rpm. The pET3c plasmid was extracted using HP Plasmid Mini Kit.
[0109] 2. Expression of Sphingomonas MlrA and preparation of MlrA crude enzyme preparation
[0110] PCR purified product and pET3c plasmid were used to construct recombinant expression vector pET3c-MlrA. The recombinant expression vector pET3c-MlrA was identified and transformed into the expression strain Escherichia coli BL21(DE3)pLysS. The resulting recombinant bacteria were expanded an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com