Normalized cDNA library of ocean medaka specific tissue and preparation method thereof
A medaka and specific technology, applied in the field of cDNA library, can solve the problems of incomplete genetic information, high proportion of copies, no product seen, etc., and achieve the effect of convenient analysis and research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Construction of a uniform cDNA library of liver tissue of marine medaka
[0043] The steps of this embodiment are as follows:
[0044] 1. Culture of Marine Medaka
[0045] The marine medaka fish comes from commercial farms in Taiwan, purchased and donated by the Department of Biology and Chemistry, City University of Hong Kong. The laboratory culture conditions of marine medaka fish are seawater dissolved oxygen level 5.0-7.0mg / L, water temperature 28±2℃, density setting 0.3-0.5L per fish, salt concentration 30mg / L, light cycle 14h lighting / 10h dark room, feeding 5 times a day, sex ratio male: female=1:1.5. Sexually mature adult fish with a body length of 3.5-4cm were selected as research objects in the experiment.
[0046] 2. Isolation of Marine Medaka Liver Tissue
[0047] Select 10 adult marine medaka fish, including 5 females and 5 males, kill them quickly by decapitating them, wash them twice with PBS buffer, separate the liver tissue aseptically, an...
Embodiment 2
[0058] Example 2 Construction of a homogeneous cDNA library of gonad tissue of marine medaka fish
[0059] Select 10 adult marine medaka fish, including 5 females and 5 males, kill them quickly by decapitating them, wash them twice with PBS buffer, separate the gonad tissue by aseptic operation, and place them in a bottle containing buffer. Wash twice, transfer to Eppendorf tubes, and collect mixed gonad tissues by centrifugation at 2000rpm / min at low speed. Other steps are the same as in Embodiment 1.
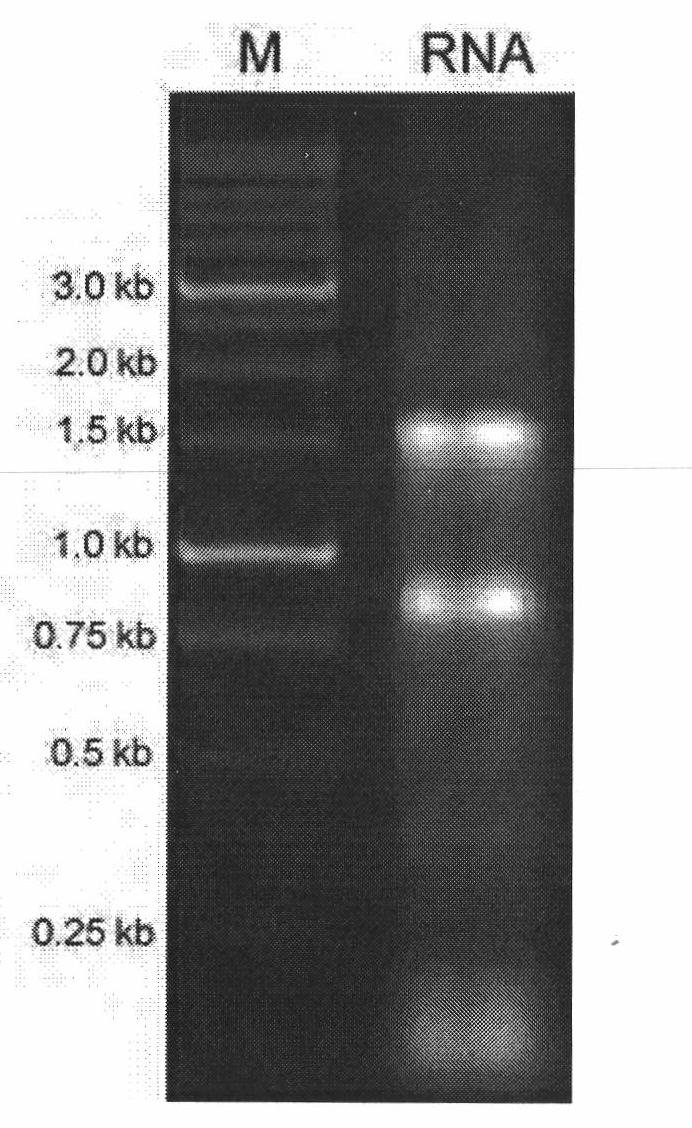
[0060] attached Figure 4 It is the result of 1% agarose gel electrophoresis of the total RNA of the gonad tissue of marine medaka fish. The 28SrRNA and 18S rRNA bands are bright and clear, and the 5S rRNA band is weak, indicating that the integrity of the total RNA is better.
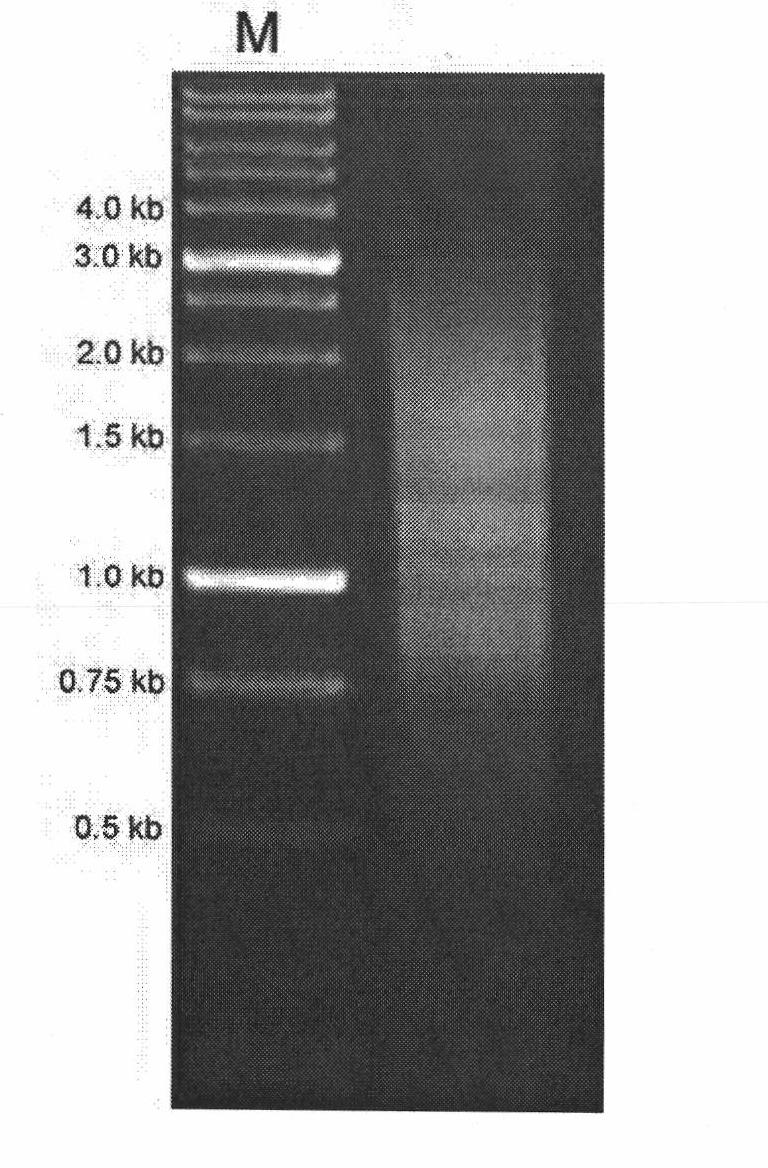
[0061] The results of 1% agarose electrophoresis before and after normalization of LD-PCR amplification products of marine medaka gonad tissue cDNA library are shown in the appendix Figure 5 And at...
Embodiment 3
[0063] Example 3 Identification of the homogenized cDNA library of liver tissue of marine medaka fish
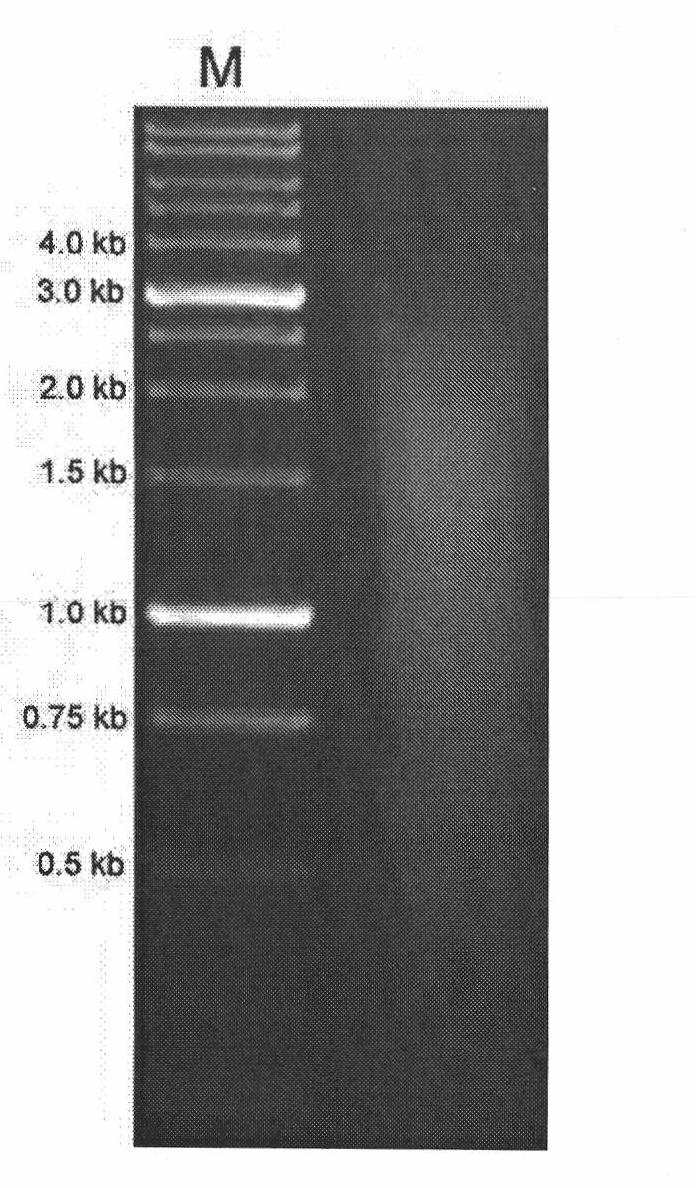
[0064] Randomly select 22 clones from the LB culture plate, inoculate them in 10ml of LB culture medium, cultivate them at 37°C for 28 hours, collect the cultures, and use the sequence primers of SEQ ID No.4 and SEQ ID No.5 to amplify the insert fragments. The increasing conditions are 95°C, 30 seconds; 65°C, 30 seconds; 72°C, 3 minutes; 30 cycles. 5ul of the product was subjected to 1% agarose gel electrophoresis, and the size of the inserted fragment was observed. The results are shown in the appendix Figure 7 , the 22 clones selected at random all have inserts, the size of which inserts ranges from 500bp-3kb, most of which are around 1kb-2kb. Sequencing at the same time, the sequencing primer sequences are SEQ ID No.4 and SEQ ID No.5, the sequencing results were compared in the gene bank, and one of the sequencing results was 99% consistent with the gene sequence gb|EF39...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com