Polymerase chain reaction (PCR)-restricted fragment length polymorphism (RFLP) method for quickly detecting single nucleotide polymorphism of goat Lhx3 gene
A PCR-RFLP, single nucleotide polymorphism technology, applied in the field of molecular genetics, can solve the problem that the goat Lhx3 gene polymorphism has not been reported, and achieve the effect of low cost and high precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
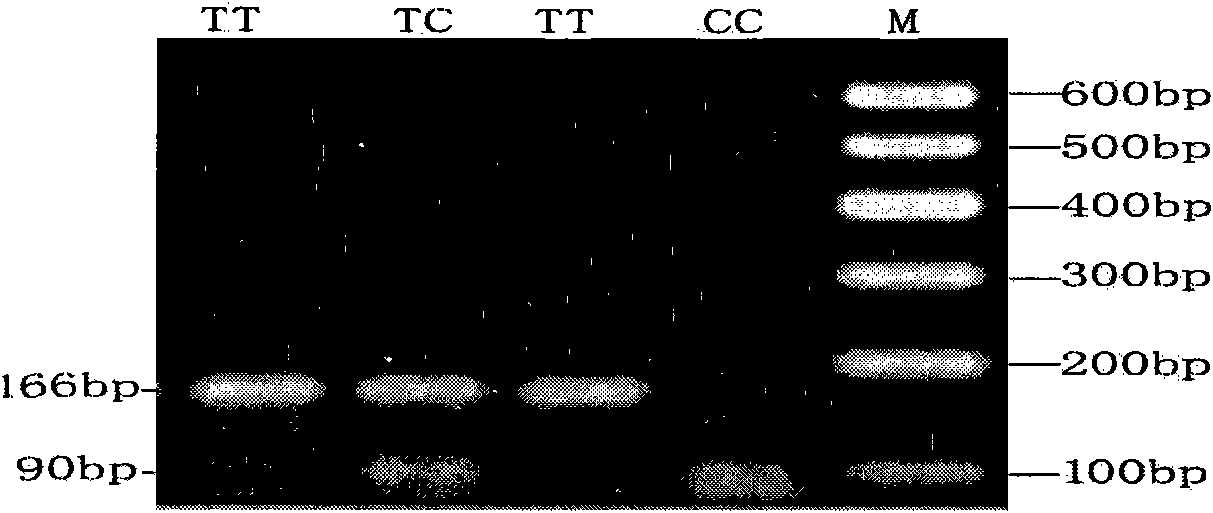
[0032] The present invention designs primers according to the conserved sequence of the Lhx3 gene, respectively uses the genomic DNA pools of four goat varieties as templates, performs PCR amplification, purifies the PCR products, and obtains the partial sequence of the Lhx3 gene of goats such as HM446543 after sequencing. The present invention will be described in detail below, which is an explanation of the present invention rather than a limitation.
[0033] I. Cloning and DNA sequencing of partial DNA sequence of goat Lhx3 gene
[0034] 1. Collection and processing of goat blood samples
[0035] Take 10 mL of goat blood, add 500 μL of 0.5 mol / L EDTA to anticoagulate, slowly invert 3 times, put it in an ice box, and store it at -80°C for later use.
[0036] The present invention adopts 4 kinds of goat breeds totaling 1073 individuals, including 2 milk goat breeds, 1 cashmere goat breed and 1 meat goat breed, specifically: (1) 229 blood samples of Guanzhong goat were collec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com