Aptamer for grouping different subtype non-small cell lung cancers and screening method thereof
A non-small cell lung cancer, nucleic acid aptamer technology, applied in biochemical equipment and methods, individual particle analysis, material excitation analysis, etc., can solve the problems of insufficient specificity and sensitivity, and achieve accurate diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1. Screening of nucleic acid aptamers for typing different subtypes of non-small cell lung cancer
[0046] 1. Design and synthesis of random nucleic acid library
[0047] Design and synthesize a random nucleic acid sequence library comprising 20 nucleotides at both ends and 45 nucleotides in the middle as follows: 5'-ACGCTCGGATGCCACTACAG(N 45 ) CTCATGGACGTGCTGGTGAC-3'.
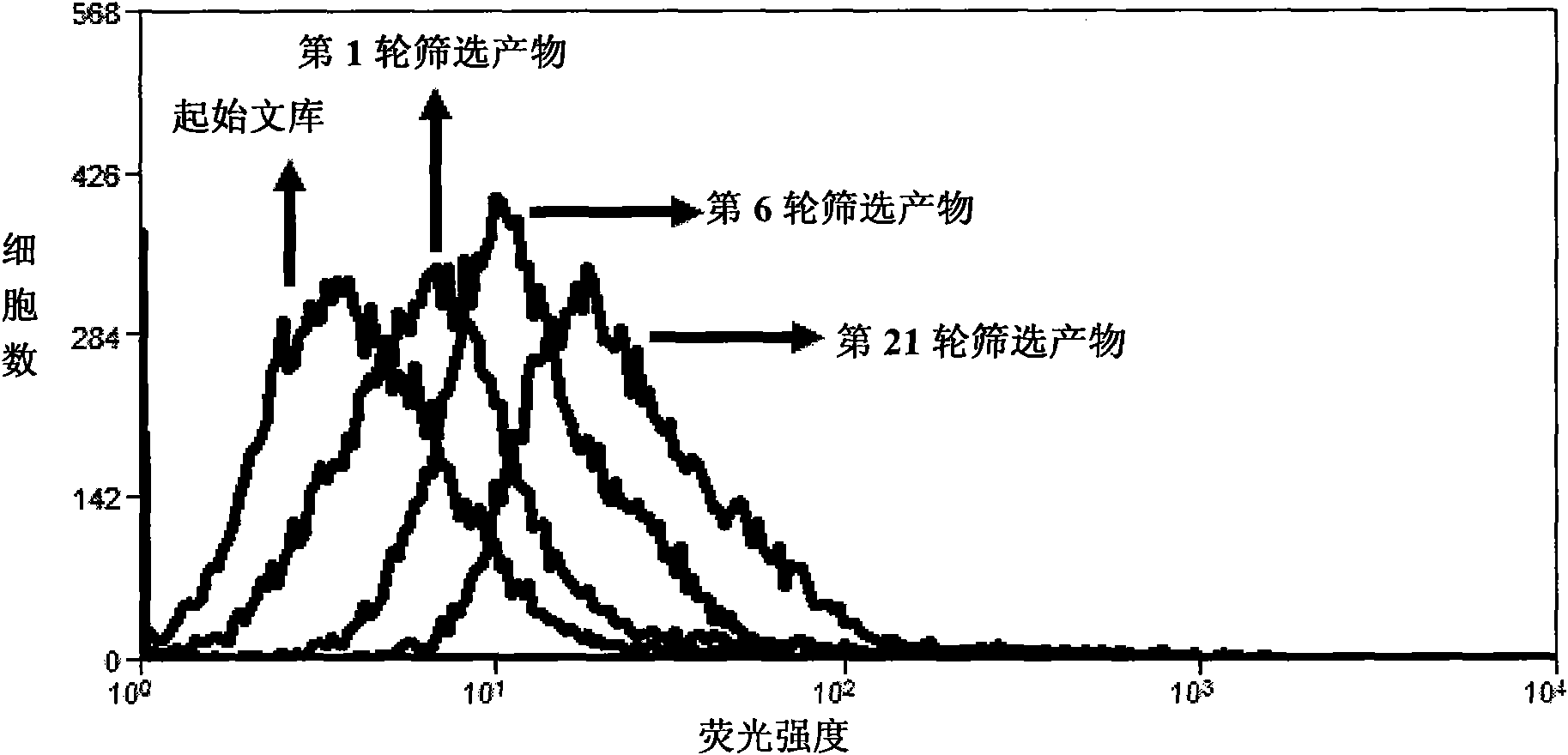
[0048] 2. Screening of nucleic acid aptamers
[0049] Dissolve the random nucleic acid library in binding buffer (PBS, 150mM NaCl, 5mM MgCl 2 , 1mg / ml yeast transfer RNA), incubated with lung adenocarcinoma cell line A549 cells on ice for 1 hour; washed with buffer solution (PBS, 150mMNaCl, 5mM MgCl 2 ) after washing, the A549 cells were scraped off, and then the DNA bound to the cell surface was dissociated by heating; the dissociated DNA was incubated with the large cell lung cancer cell line HLAMP cells on ice for 1 hour, and the supernatant was collected and used DNA was used as a templa...
Embodiment 2
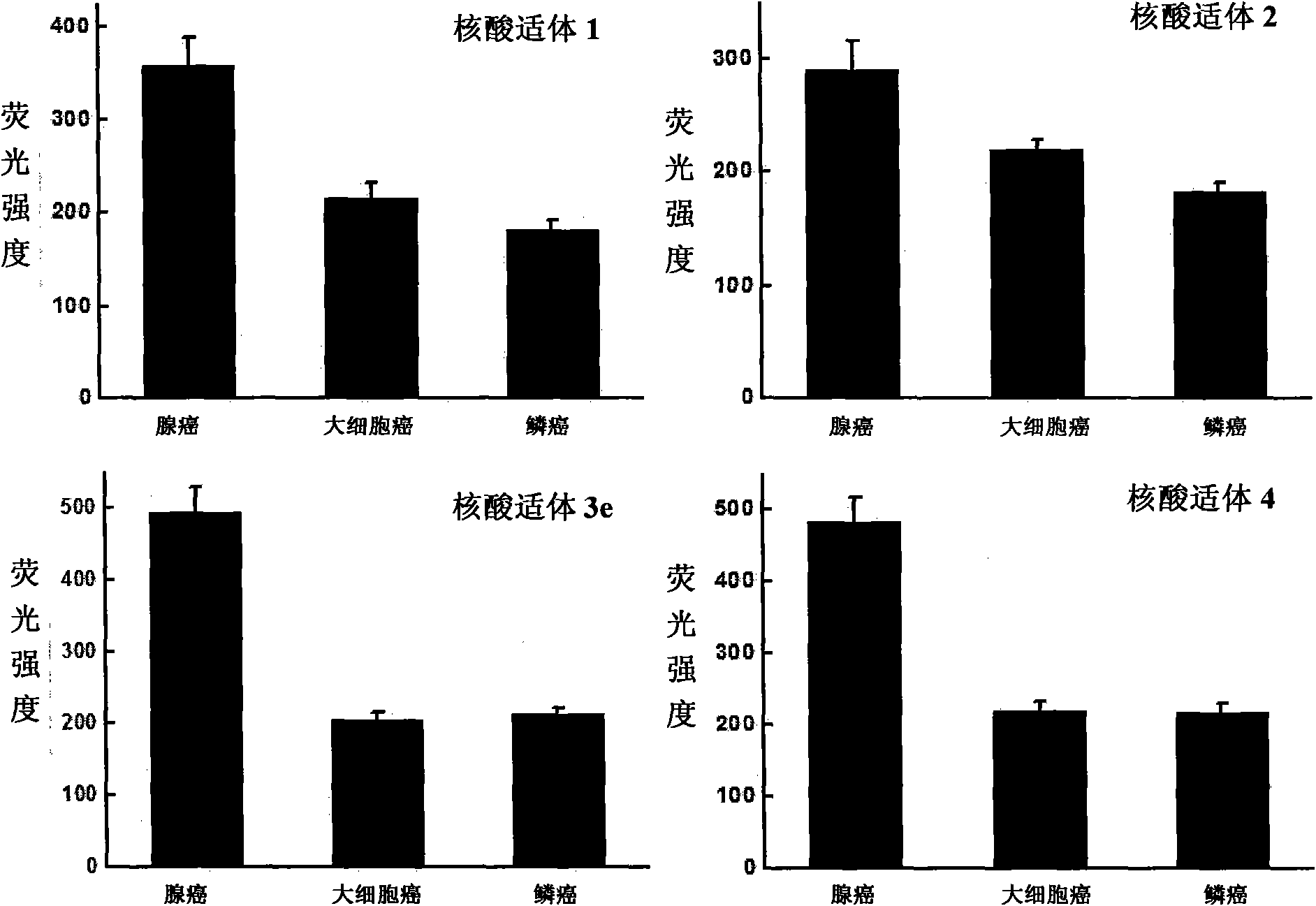
[0062] Example 2. Applying nucleic acid aptamers to type different subtypes of small cell lung cancer
[0063] The four aptamers obtained in Example 1 were labeled with fluorescein isothiocyanate (FITC). The random nucleic acid library of Example 1 was labeled with fluorescein isothiocyanate (FITC).
[0064] Different cultured tumor cells were washed twice with PBS, then treated with 0.02% EDTA for 3 minutes, and washed twice with PBS.
[0065] Each tumor cell was subjected to the following four groups of treatments, and each treatment was repeated three times, and the results were averaged:
[0066] Treatment 1: by cell counting, about 300,000 cells were respectively dispersed in the binding buffer solution, and then fluorescein isothiocyanate (FITC) was added to label the nucleic acid aptamer 1 (the final concentration was 100 nM).
[0067] Treatment 2: by cell counting, about 300,000 cells were collected and dispersed in the binding buffer solution, and then fluorescein i...
Embodiment 3
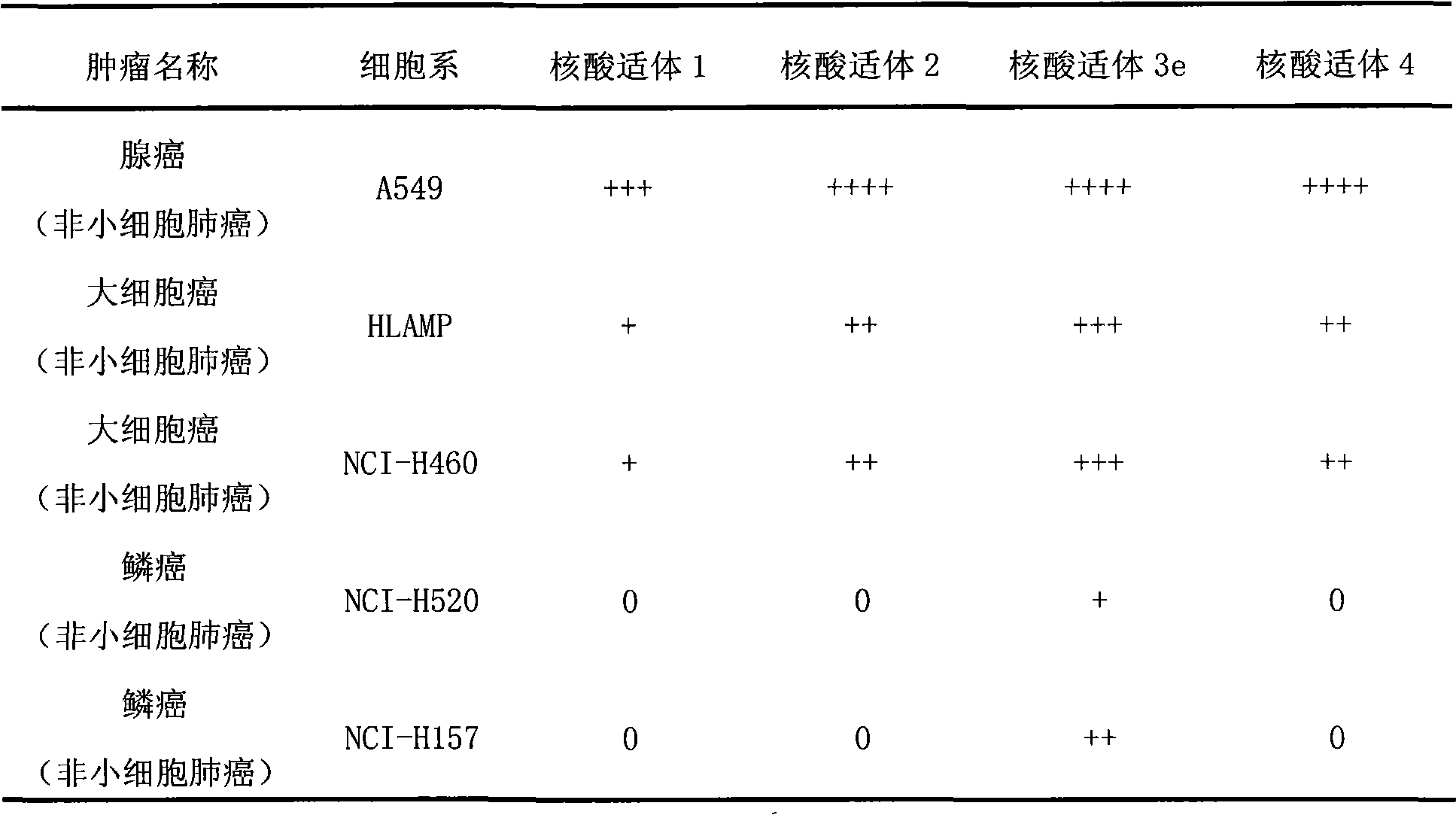
[0076] Example 3. Detection of the binding of the nucleic acid aptamers listed in Table 1 to lung cancer tissue sections
[0077] Formaldehyde-fixed paraffin-embedded tissue sections from different volunteers with lung cancer were dewaxed twice in xylene at room temperature for 20 minutes each time, and then immersed in a series of ethanol for hydration (100%, twice, 1 minute each time) ; 95% ethanol, 1 time, 1 minute; 70% ethanol, 1 time, 1 minute). The hydrated tissue sections were then immersed in buffer solution and heated at 95°C for 15 minutes. After the tissue sections returned to room temperature, they were incubated with binding buffer containing 20% fetal bovine serum and calf thymus DNA for 1 hour at room temperature. After washing, the tissue sections were respectively combined with tetramethylrhodin-labeled nucleic acid aptamers (200nM) Incubate in buffer for 1 hour at room temperature. After staining, the tissue sections were washed 3 times with washing buffe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com