Separation, cloning and identification of promoters of porcine citric dehydrogenase genes IDH3beta
A technology of isocitrate dehydrogenase and promoter, applied in the field of pig breeding and molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1: Preparation of the upstream 5' flanking sequence of porcine citrate dehydrogenase gene IDH3β gene
[0074] The TAIL-PCR method (Liu et al., 1995) was used to isolate the promoter sequence of porcine citrate dehydrogenase gene IDH3β. This method required two rounds of PCR amplification for each step. The PCR reaction conditions are shown in Table 1.
[0075] Table 1. Three rounds of amplification conditions for TAIL-PCR
[0076]
[0077]
[0078] The reaction system is as follows: (1) First round: 100ng porcine genomic DNA, 0.2μM gene-specific primer 1 (SE), 2μM random primer R (R1, R2, R3 and R4), 2.5μL 2mM dNTP, 1U Taq DNA polymerase , 2.5 μL of 10×Taq DNA polymerase buffer, added to a total volume of 25 μL; (2) the second round. Dilute the first-round PCR product 50 times, take 1 μL as template, 0.2 μM SE gene-specific primer 2 (SF), 2 μM random primer R (R1, R2, R3 and R4), 2.5 μL 2mM dNTP, 1U Taq DNA polymerase , 2.5 μL 10×Taq DNA polymerase, adde...
Embodiment 2
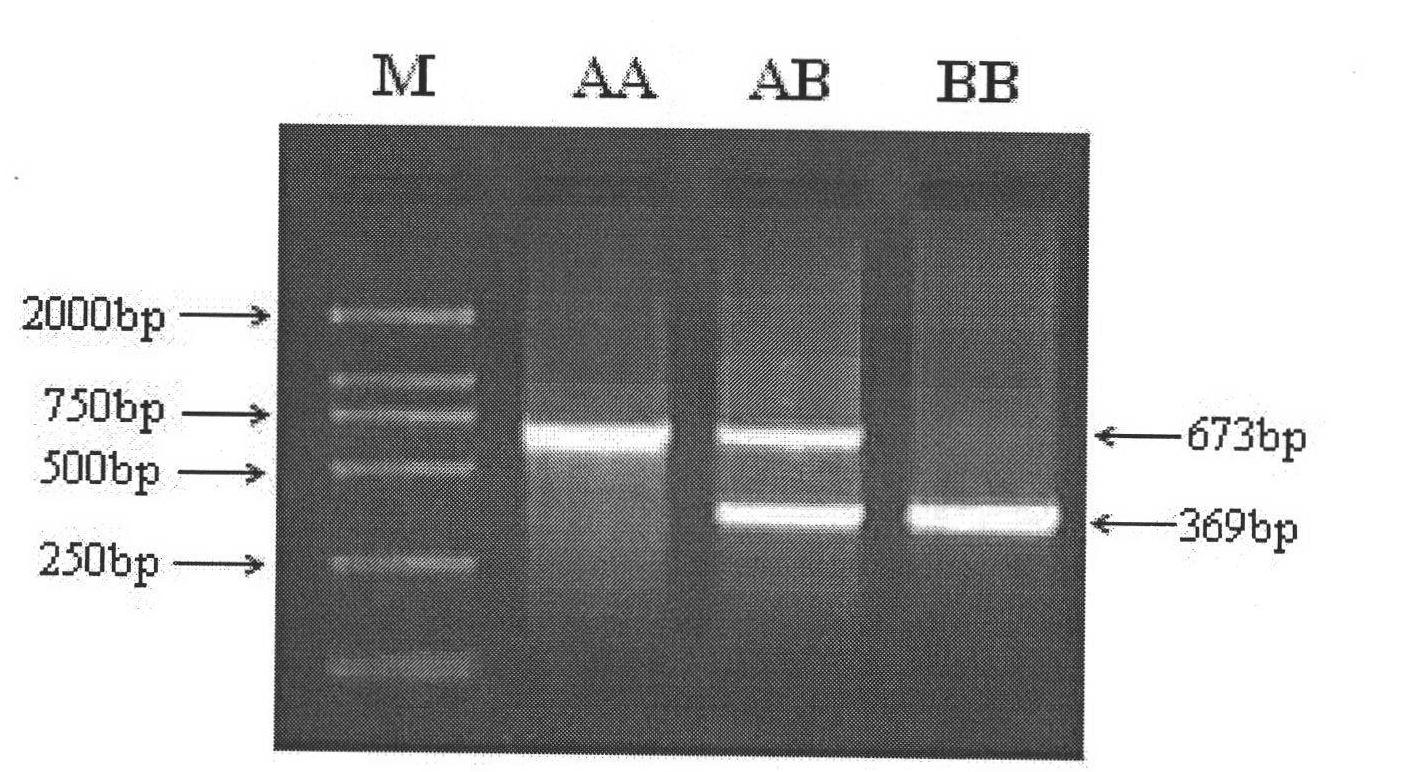
[0095] Example 2: Detection method for the mutation of the upstream 5' flanking sequence of porcine citrate dehydrogenase gene IDH3β
[0096] Genomic DNA was extracted from 2 foreign blood-related pig breeds "Large White Pig" and 2 Chinese native pig breeds "Meishan Pig", and the 2447bp 5' flanking sequence of porcine citrate dehydrogenase gene IDH3β obtained according to Example 1 was used as the test material , design the following primers, the primer numbers and nucleotide sequences are as follows:
[0097] Forward primer 31: TGGCTCTGTCCAAGGGTT;
[0098] Reverse primer 32: AACTGGGATTTGTACTGC.
[0099] Use primers 31 and 32 to carry out PCR amplification in the genomic DNA of the above two large white pigs and two Meishan pigs. The PCR reaction system is 25 μl, in which the template DNA is 50 ng, the concentration of dNTPs is 200 μmol / L, and the concentration of each primer is 0.3 μmol / L, 1U Taq DNA polymerase (Biostar International, Canada), add deionized water to a tota...
Embodiment 3
[0101] Example 3: Construction of expression vector containing porcine citrate dehydrogenase gene IDH3β upstream 5' flanking sequence
[0102] 1. Primer Design
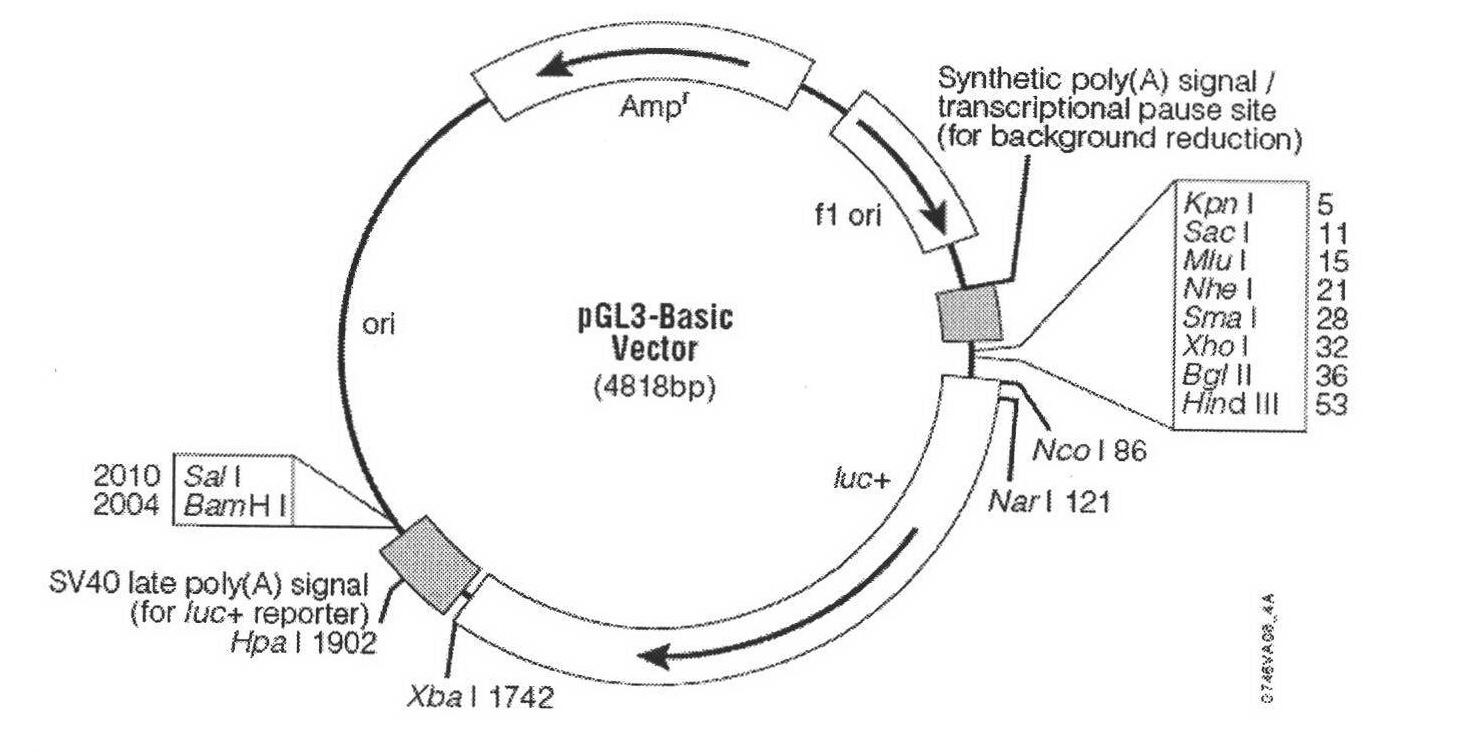
[0103] According to the characteristics of the multiple cloning site of the pGL3-Basic vector (purchased from Promega, USA) and the promoter sequence of the porcine ACL gene, primers were designed using the deletion method, and there were XhoI and MluI restriction sites at the 5' ends of the upstream and downstream primers, respectively. (The sequence of the restriction site is underlined, as shown in Table 3), and three protective bases are respectively added upstream of the recognition sequence of the restriction site to ensure that the restriction site is not lost. The large white pig genomic DNA was used as a template for PCR reaction, and the annealing temperature of PCR amplification was 58°C. The primer numbers and their nucleotide sequences are as follows:
[0104] Forward primer 11: GCCACGCGTGAGAGAGACGCTTTC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com