Method for improving yield of Streptomyces antibiotics and plasmid thereof
A technology of streptomyces and antibiotics, applied in the field of biomedicine, can solve the problems of limited influence of metabolites and insufficient metabolic engineering, and achieve significant industrial application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Preparation of a single-component mutant strain producing candicidin D
[0049] (1) Construction of plasmids (used to transfer into hosts and undergo homologous recombination with chromosomes to obtain target mutations)
[0050] (1) Plasmid construction for KR21 functional domain mutation:
[0051] The 3615-bp EcoRI-KpnI DNA fragment was recovered after EcoRI and KpnI digestion of the Streptomyces FR-008 library cosmid pHZ220 as a PCR template, primers P1 and P3 were used to amplify the 528-bp DNA fragment; P4 and P2 were used to amplify the 618 - bp DNA fragments. There is an 18-bp overlap between P3 and P4, and a mutation site (Y1526F) is introduced in the overlapping region, and a DNA restriction enzyme site (ApoI) is introduced at the same time for the screening of the mutation (such as figure 1 shown). A 1146-bp DNA fragment was obtained by the second PCR (primers were P1 and P2; the template was 1 / 100 of the two PCR products recovered from the agaros...
Embodiment 2
[0094] Firstly, the homologous recombination plasmid pJTU572 was transformed into Streptomyces FR-008 cells by conjugative transfer to undergo homologous recombination, and the DH18 mutant was screened and obtained. Then the plasmid pJTU573 was transferred into the cells of the DH18 mutant strain to mutate KR21 through homologous recombination, and the KR21 and DH18 double mutant strain ZYJ-6 was finally screened.
Embodiment 3
[0095] The improvement of embodiment 3 streptomyces antibiotic output
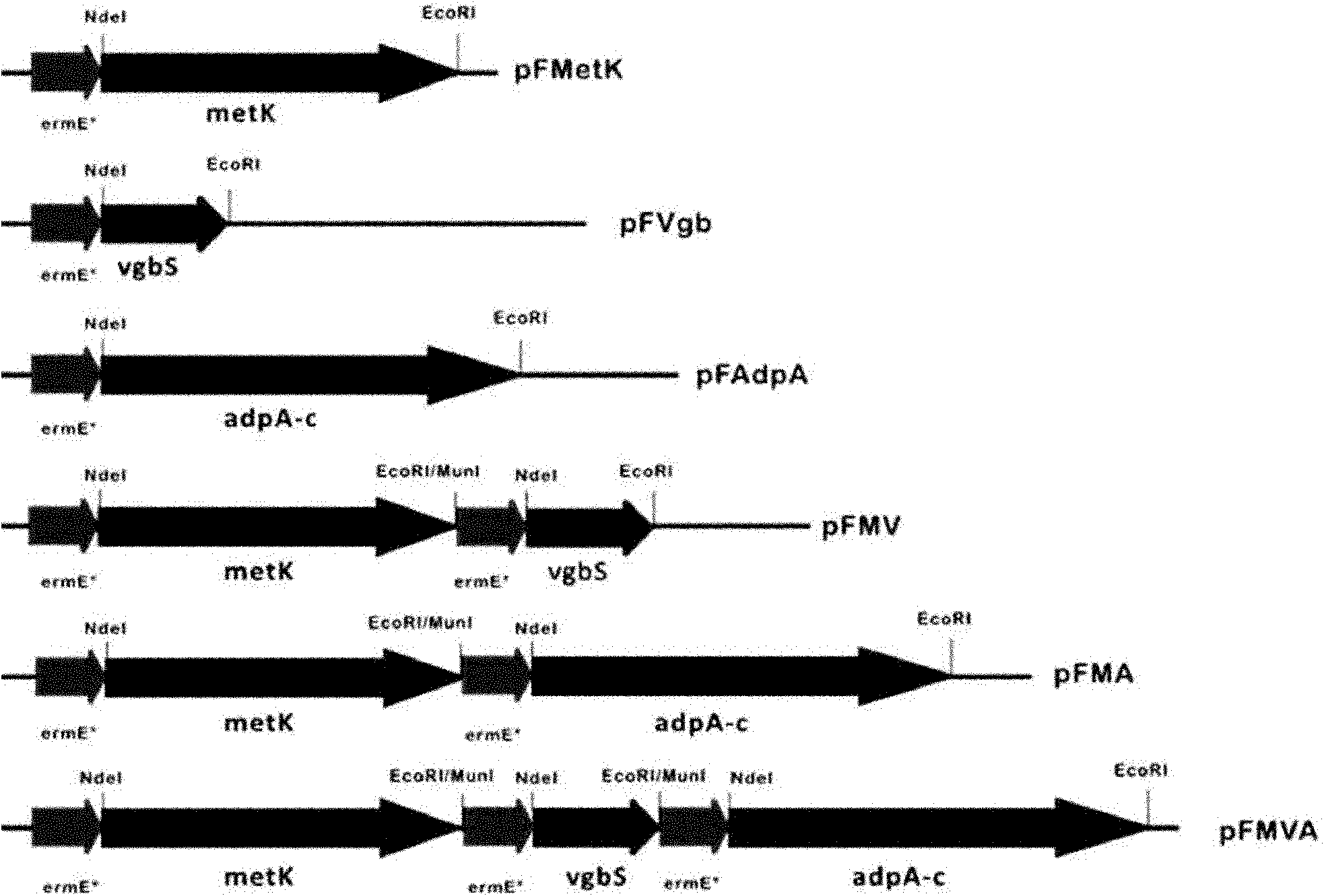
[0096] Step 1, construction of expression plasmid.
[0097] The gene expression vector pIB139 involved in this example is a strong promoter PermE containing the erythromycin resistance gene. *The Streptomyces integrative vector can be transferred into Streptomyces through biparental conjugation between Escherichia coli and Streptomyces. After site-specific recombination, it is integrated on the chromosome of Streptomyces, and is replicated and inherited along with the chromosome. figure 1 Maps were constructed for all expression plasmids involved in this example.
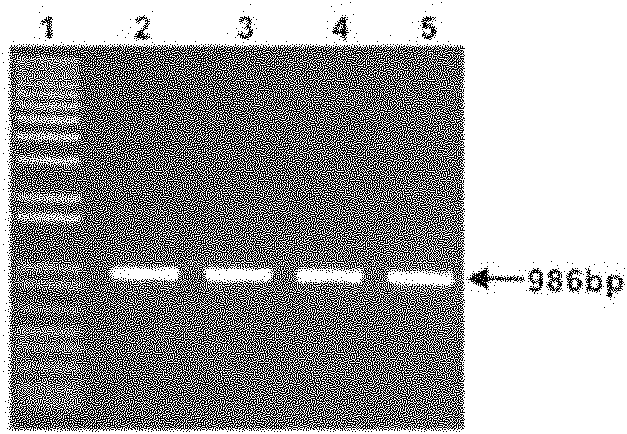
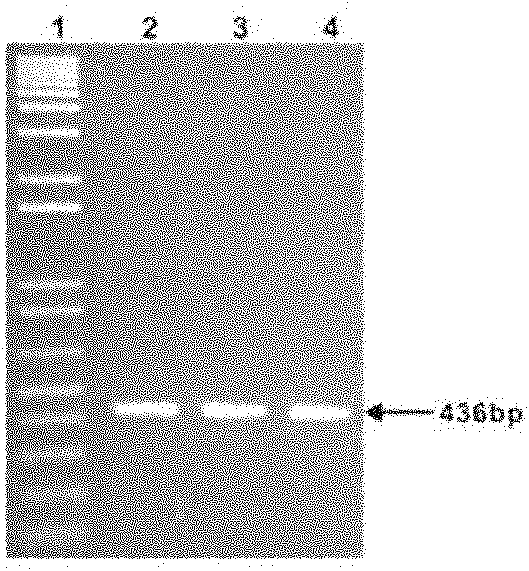
[0098] Step 2, transfer the plasmid into the Streptomyces host and screen and verify the conjugative transfer
[0099] Transform the constructed plasmid into Escherichia coli ET12567 / pUZ8002, pick the transformants and cultivate the Escherichia coli ET12567 / pUZ8002 carrying the transferred plasmid in liquid LB medium, where there is chlorine ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com