RV (rabies virus) dominant-epitope peptide antigen and application thereof
A rabies virus and dominant epitope technology, applied in the field of rabies vaccine, can solve the problems of low sensitivity, high false negative rate, poor specificity, etc., and achieve the effect of improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Determination of the dominant epitope antigen segment of rabies virus
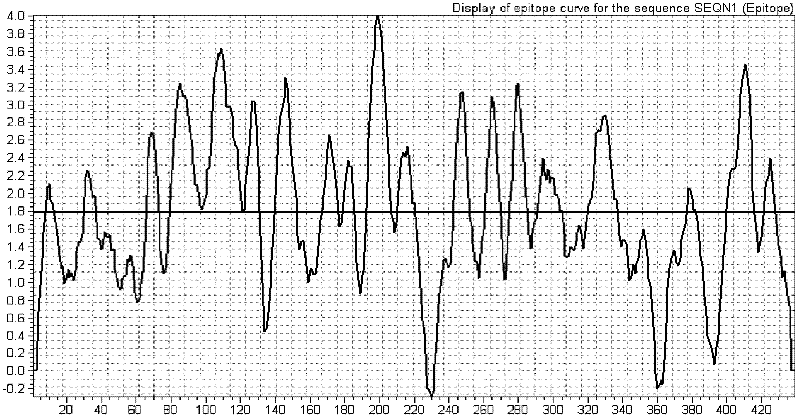
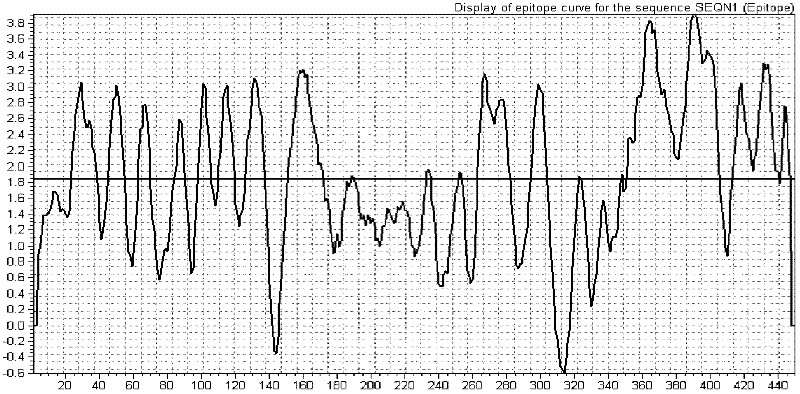
[0032] Using BIOSUN biological analysis software to analyze the epitope distribution of rabies virus G protein and N protein, first input their amino acid sequence, and then search for B cell epitope, the obtained epitope distribution map is as follows figure 1 with figure 2 shown. Determine the specific dominant epitope antigen segment according to the epitope distribution, which are: the dominant epitope segment GP-1 (1-200aa), the amino acid sequence is SEQ ID NO: 1; the dominant epitope segment GP-2 ( 301-400aa), the amino acid sequence is SEQ ID NO: 2; the dominant epitope segment GP-3 (60-230aa), the amino acid sequence is SEQ ID NO: 3; the dominant epitope segment GP-4 (231-345aa) , the amino acid sequence is SEQ ID NO: 4; the dominant epitope segment NP-1 (301-420aa), the amino acid sequence is SEQ ID NO: 5; the dominant epitope segment NP-2 (20-180aa), the amino acid sequence ...
Embodiment 2
[0033] Example 2: Cloning of rabies virus dominant epitope peptide antigen coding gene
[0034] 1. Primer design
[0035] According to the full-length nucleotide sequences of G protein and N protein and the nucleotide sequences of 8 dominant epitope peptide antigens, the upstream and downstream primers were designed and synthesized respectively, and the restriction enzymes used were Xho I and Xba respectively. I, BamH I or EcoR I.
[0036] The primers used to amplify the full-length G protein gene are GF and GR, and the nucleotide sequences are shown in SEQ ID NO: 9 and SEQ ID NO: 10, respectively;
[0037] The primers used to amplify the full-length gene of N protein are NF and NR, and the nucleotide sequences are shown in SEQ ID NO: 11 and SEQ ID NO: 12, respectively;
[0038] The primers used to amplify the gene encoding the dominant epitope segment GP-1 are GF-1 and GR-200, and the nucleotide sequences are shown in SEQ ID NO: 13 and SEQ ID NO: 14, respectively;
[0039]...
Embodiment 3
[0052] Example 3: Expression and purification of rabies virus dominant epitope peptide antigen
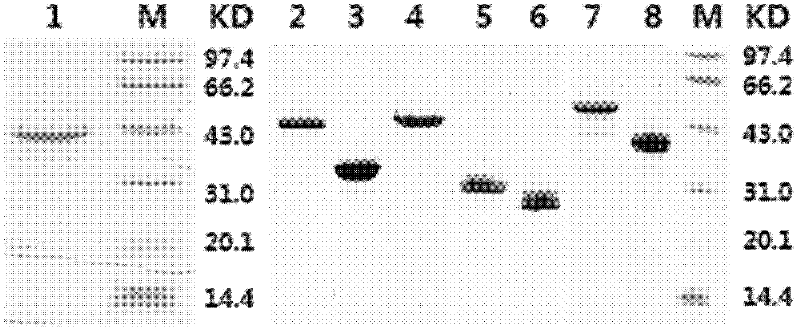
[0053] Transform the Escherichia coli strain with the obtained recombinant plasmid, heat-induced expression, collect the thalline, weigh the wet weight of the precipitate, suspend the precipitate with 10 times the volume of 20mmol / L pH8.0TE buffer, add lysozyme (1mg / ml suspension), stirred magnetically for 10 minutes at room temperature. Sonicate the bacteria in an ice bath for 30 seconds each time, with an interval of 30 seconds, a total of 10 times. Centrifuge at 1,2000 rpm at 8°C for 20 minutes, discard the supernatant, wash the precipitate once with 1 mol / L NaCl (prepared with TE), wash twice with TE, and collect the precipitate. Dissolve the precipitate with a dissolving solution (25mM Tirs-HCl, 1% β-mercaptoethanol, 8M urea, pH 7.8), put it on a Ni-Sepharose Fast Flow column, and use 3 column volumes of binding buffer (20mM Tirs-HCl , 0.1% β-mercaptoethanol, 6M urea, pH 7.8...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com