Sugarcane smut bacteria nest type polymerase chain reaction (PCR) quick detection method
A sugarcane smut and detection method technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganism measurement/inspection, etc., can solve the problem of being easily interfered by other pathogens, unable to accurately detect, and poor detection specificity and other problems, to achieve the effect of fast detection, high accuracy and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
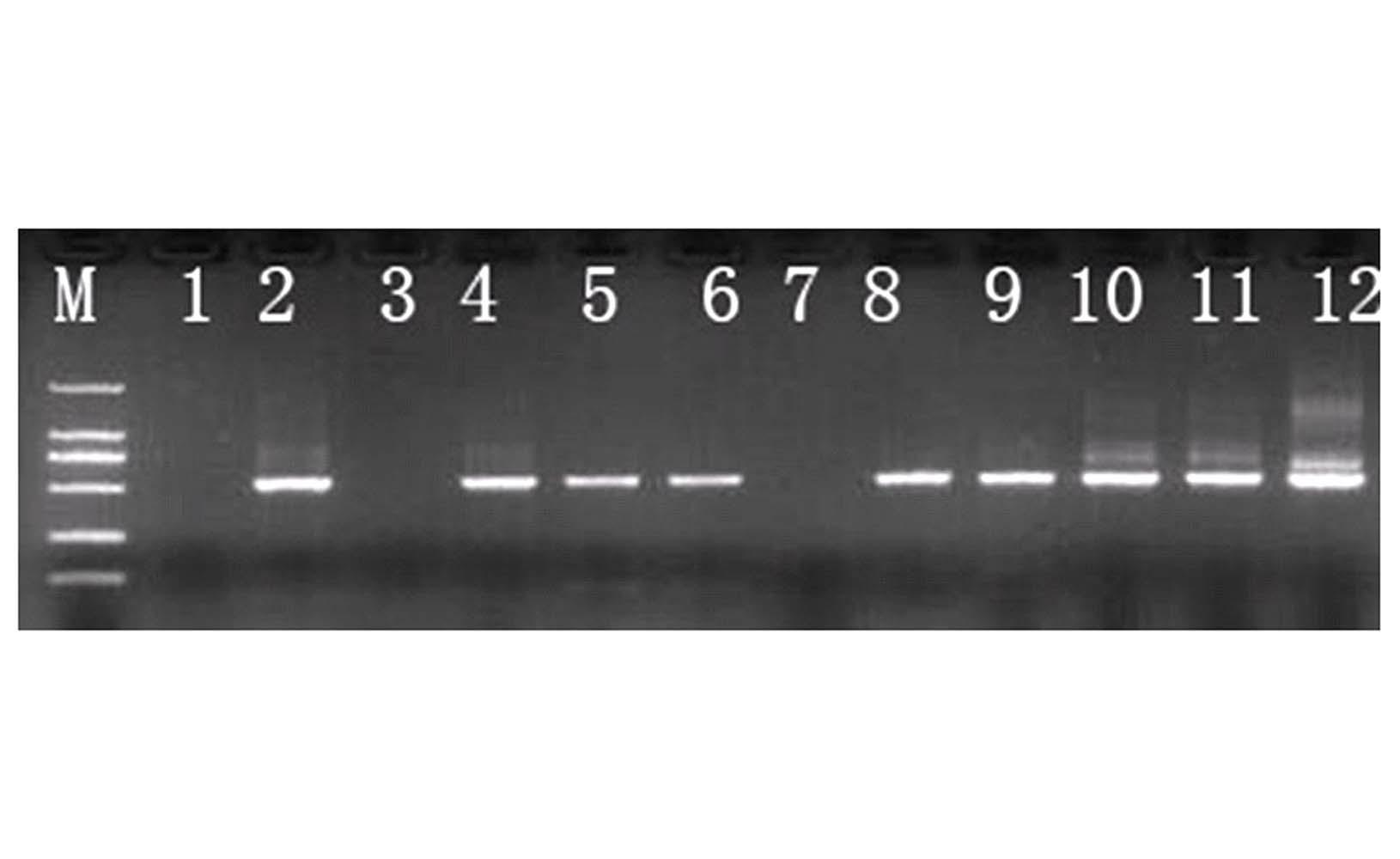
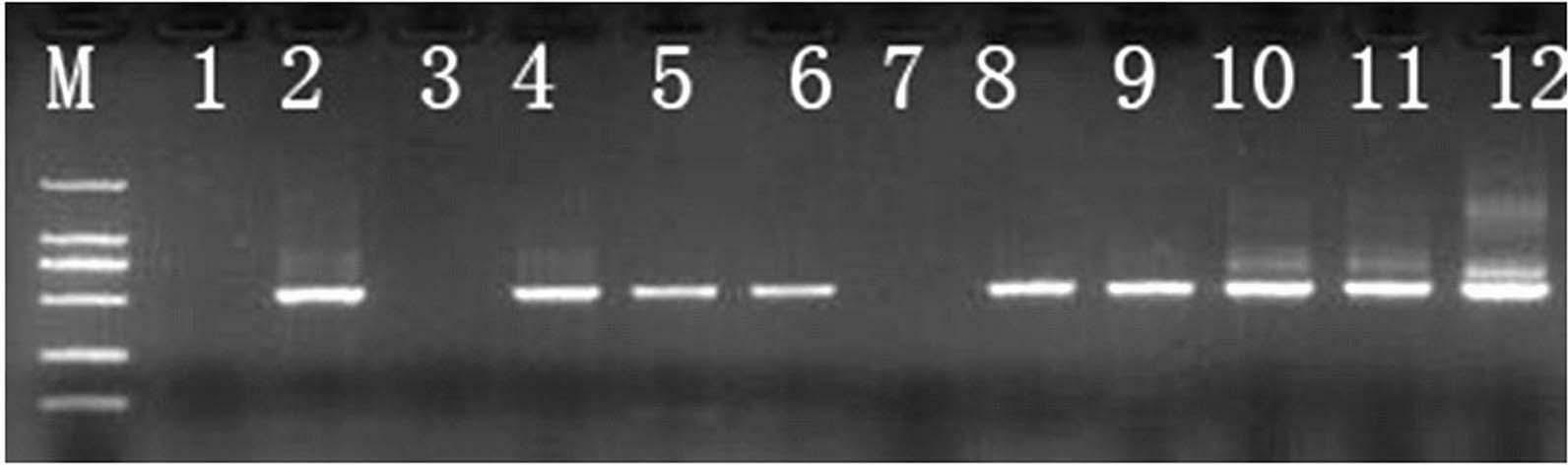
[0026] The nested PCR rapid detection method of sugarcane smut comprises the following steps:
[0027] Step A, genomic DNA extraction:
[0028] In April 2011, 9 heart leaves of sugarcane variety Xintaitang No. 22 seedlings artificially inoculated with teliospores of sugarcane smut were randomly collected (without early symptoms of sugarcane smut). Genomic DNA was extracted, and the samples were obtained and numbered as samples 4-12 in sequence;
[0029] Step B, the first round of PCR amplification:
[0030] Using samples 4-12 as templates, the first round of PCR amplification products were obtained through PCR amplification using universal primers ITS4 and ITS5 of the fungal ribosomal gene transcription spacer ITS;
[0031] The primer sequence is: ITS4:5`-TCCTCCGCTTATTGATATGC-3`;
[0032] ITS5:5`-GGAAGTAAAAGTCGTAACAAGG-3`;
[0033] PCR reaction system: Genomic DNA 1 μL, 5 μmol / L ITS4 and ITS5 primers 1 μL each, Taq enzyme 0.2 μL (5U / μL), 10×PCR reaction buffer (containin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com