Methylated DNA Detection Method Based on Endonuclease Digestion
An endonuclease and detection method technology, applied in the field of detection of methylated DNA, can solve the problems of inapplicable mixed samples, large sample size, and complicated methods, and achieve the goal of not being easily disturbed, requiring small sample sizes, and methods simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach ( 1
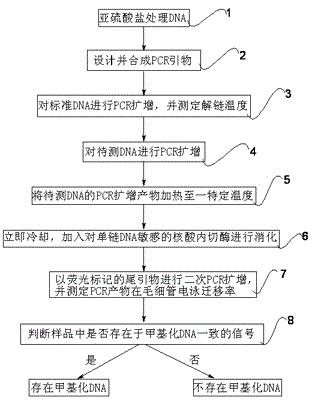
[0031] Step 1, Treat with sulfite and standard unmethylated DNA with standard methylated DNA.
[0032] Wherein, the sulfite is specifically sodium sulfite; the DNA to be tested treated with sulfite can be specifically a human genome gene. The treatment steps of the sulfite treatment of the DNA to be tested are as follows: denature the DNA to be tested with 0.3M NaOH, add a mixed solution containing 5M sodium sulfite and 0.5mM hydroquinone with a pH value of 5.0, and incubate at 60°C for 4 hours in the dark; desulfurization and purify the DNA. The unmethylated cytosine (C) in the DNA to be tested is converted into uracil (U) through the above treatment process, and the methylated cytosine (C) remains unchanged.
[0033] Wherein said standard unmethylated DNA and standard methylated DNA refer to known pure unmethylated DNA and methylated DNA.
[0034] Step 2, designing and synthesizing PCR primers.
[0035] In the target region to be detected, select a sequence rich in CpG si...
Embodiment approach ( 2
[0054] On the basis of embodiment (1), DNA extracted from actual clinical samples was used as the sample to be tested, and the practicability of the present invention was tested under the same conditions as the above embodiment (1).
[0055] Among them, the same conditions as in the above-mentioned embodiment (1) mean that all operations are the same as the above-mentioned embodiment (1) except that the DNA extracted from the actual clinical sample is used as the sample to be tested.
[0056] The clinically extracted DNA samples to be tested are human normal and cancer cell DNA, tumor tissue genomic DNA, blood cell DNA, serum DNA, various body fluid DNA or various excrement DNA samples.
[0057] The cancer cells can be lung cancer cells, gastric cancer cells, colon cancer cells, rectal cancer cells, liver cancer cells, breast cancer cells, lymphoma cells, bladder cancer cells, ovarian cancer cells, kidney cancer cells or pancreatic cancer cells; The tissue can be lung cancer t...
Embodiment approach ( 3
[0059] On the basis of the implementation methods (1) and (2), taking the sequence of the transcription promoter region of the P16 gene as an example, the specific PCR primers are designed as follows:
[0060] Methylation of the transcription promoter region of P16 gene is a characteristic of many cancer cells such as lung cancer. The sequence of the transcription promoter region of the P16 gene is as follows:
[0061] Homosapiensp16protein (CDKN2A) gene, CpGislandandpartialcds, DQ325544.1
[0062] CGGACCGCGTGCGCTCGGCGGCTGCGGAGAGGGGGAGAGCAGGCAGCGGGCGGCGGGGAGCAGCATGGAGCGGGCGGCGGGGAGCAGCATGGAGCCTTCGGCTGACTGGCTGGCCACGGCCGCGGCCCGGGCTCGGGTAGAGGAGGTGCGGGCGCTGCTGGAGGCGGGGCGCTGCCCAACGCACCGAATAGGATTACGCCG
[0063] Methylated DNA sequence, the underlined part is the region to be detected;
[0064] CGGATCGCGTGCGTTCGGCG GTTGCGGAGAGGGGGAGAGTAGGTAGCGGGCGGCGGGGAGTAGTATGGAGCGGGCGG CGGGGAGTAGTATGGAGTTTTCGGTTGATTGGTTGGTTACGGTCGCGGTTCGGGTTCGGGTAGAGGAGGTGCGGGCG TTGTTGGAGGCGGGGGCGTTGTTTAA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com