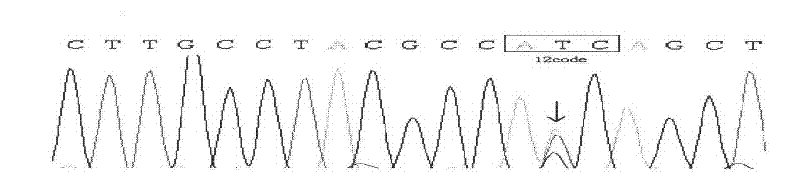Method and kit for detecting mutation of K-ras gene
A kit and gene technology, applied in the field of molecular biology, can solve the problems of high false positives, increased risk of adverse reactions and treatment costs, and the inability of K-ras mutant patients to benefit, so as to shorten the detection time and reduce the cost. , the effect of the simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Preparation of K-ras wild-type and mutant plasmid DNA, the specific steps are as follows:
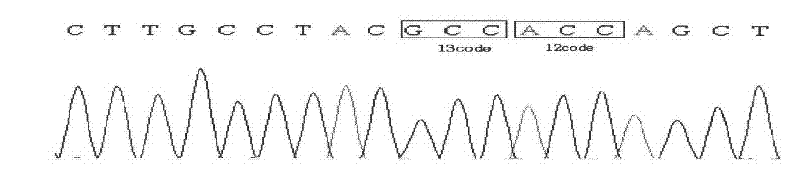
[0028] 1) Use conventional methods to extract genomic DNA from human whole blood samples. DNA sequencing proves that the samples are wild-type k-ras. The sequencing results are as follows figure 1 Shown.
[0029] 2) Design and prepare the primers for the three point mutation templates at codons 12 and 13 of the k-ras gene: primer A pair and primer pair B, primer pair C and primer pair D, primer pair E and primer pair F, The sequence is as follows:
[0030] Among them, the primer pair A includes the forward primer T1k-rasmut and the reverse primer T3k-rasmut, which amplify the upstream primer of the mutation template; the primer pair B includes the forward primer T4k-rasmut and the reverse primer T2k-rasmut, and the downstream primer of the amplified mutation template Primers; primer pair A and primer pair B amplify the mutation template of codon k-ras12 GGT-GTT.
[0031] Amo...
Embodiment 2
[0042] Embodiment 2: Detection of k-ras wild-type samples and k-ras mutant samples.
[0043] 1) Use conventional methods to extract genomic DNA from human whole blood samples. DNA sequencing proves that the samples are wild-type k-ras. The sequencing results are as follows figure 1 As shown, this was used as the K-ras wild-type sample.
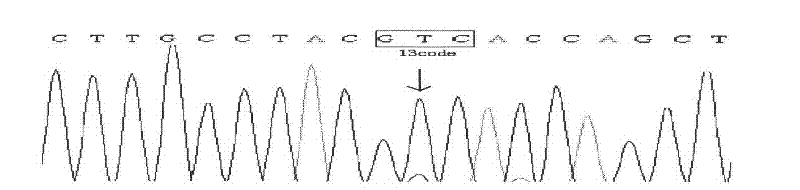
[0044] 2) DNA is extracted from tumor tissue, and DNA sequencing has at least one of the three hotspot mutations. The DNA sequencing proved to be K-ras mutant. See the sequencing results figure 2 , image 3 , Use this as a K-ras mutant sample.
[0045] 3) Use the three positive plasmid templates obtained in the examples to test the sensitivity and specificity of the kit and its specific primers. The kit includes: conventional PCR components and specific primers; the specificity includes three A forward primer and a reverse primer, wherein the sequences of the three forward primers respectively have the nucleotide sequence shown in SEQ ID No. 1, SE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com