Application of epigallocatechin-3-gallate in preparation of antitumor drug
A technology of epigallocatechin and gallate, applied in the fields of medicine and genetic engineering, can solve problems such as EGCG that have not yet been seen, and achieve the effects of not being resistant to drug resistance and inhibiting the growth of liver cancer cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 Virtual Screening of CYPJ Small Molecule Inhibitors
[0056] The present invention retrieves the X-ray diffraction crystal structure of human CYPA protein (PDB code: 1CWA) through the PDB protein structure database. This structure is the complex crystal structure of CYPA and its natural inhibitor cyclosporin A (CsA). From this structure, the active site of CYPA was determined, and several key amino acid sites in the active site that could be inhibited by CsA were determined.
[0057] In this example, for the active site of CYPJ (PDB code of CYPJ protein crystal: 1XYH; provided by Shanghai Institute of Organic Chemistry), according to the molecular docking method, several small molecule databases were virtual screened, and the binding free energy of AutoDock Vina was used to The binding affinities of the relevant small molecules were ranked (see Table 1). It was found that epigallocatechin gallate (EGCG) ranked higher with CYPA and CYPJ, and its affinity for ...
Embodiment 2
[0061] Example 2 Validation of virtual screening results using molecular docking
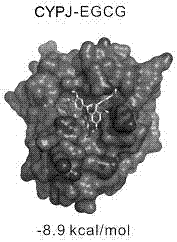
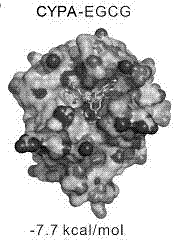
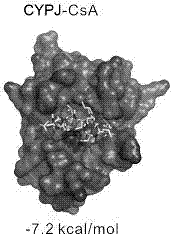
[0062] Calculate the binding free energy of EGCG, CsA and CYPA, CYPJ AutoDock Vina. Such as Figure 1-Figure 4 As shown, both EGCG and CsA can be combined at the active centers of CYPA and CYPJ, and some groups are inserted into the pockets of the active centers. Figure 4 The structure of the CsA-CYPA complex calculated by AutoDock Vina docking is consistent with the crystal structure (1CWA), indicating that the molecular docking method of the present invention is reliable. The binding energy of EGCG and CYPJ is -8.9 kcal / mol; the binding energy of EGCG and CYPA is -7.7 kcal / mol; the binding energy of CsA and CYPJ is -7.2 kcal / mol; the binding energy of CsA and CYPA is -7.3 kcal / mol mol. The above results show that EGCG can bind to CYPA and CYPJ, and the binding ability to CYPJ is stronger. It is worth mentioning that although CsA can also bind to CYPA and CYPJ, it has no higher selectivi...
Embodiment 3
[0064] Example 3 Validation of virtual screening results using BIACORE Molecular Interaction Instrument
[0065] The BIACORE Molecular Interaction Instrument is based on surface plasmon resonance technology to track the interaction between biomolecules without any markers, thus ensuring the authenticity of the experimental results to the greatest extent. During the experiment, the target biomolecules (CYPA, CYPJ proteins) were immobilized on the surface of the sensor chip, and then the small molecule compounds were dissolved in a solvent and flowed over the surface of the chip. The monitor can track the changes in the whole process of binding and dissociation between molecules in the detection solution and target biomolecules on the chip surface in real time. Binding data via BIACORE ( Figure 5-8 ), finally calculated the affinity dissociation constant KD value of EGCG and CYPA of the present invention to be 3.82×10 -7 M, the KD value with CYPJ is 6.80×10 -7 M; The KD...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com