Real-time fluorescent quantitative PCR detection method of human breast cancer genotype
A real-time fluorescence quantitative, human breast cancer technology, applied in the field of physiological and biochemical detection, can solve the problems of non-specific amplification, PCR can not quantify and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0124] Example 1 Construction of real-time quantitative standards
[0125] Using cell culture and T-A cloning techniques, synthetic primers were designed on the ERα, PR, HER-2, 18S rRNA gene sequences, and after PCR amplification, the specific products were ligated into the pGEM-19T vector. Eco R V digestion identification and sequencing analysis to obtain ERα (SEQ ID 13), PR (SEQ ID 14), HER-2 (SEQ ID 15), 18S rRNA (SEQ ID 16) plasmids, quantify the plasmid concentration, and determine the plasmid copy number. The process was repeatedly optimized in real-time quantitative qPCR to obtain standard products.
Embodiment 2
[0126] Example 2 Design of specific primers
[0127] The NCBI GenBank database was searched, the designed primers were aimed at different exons, and 18S rRNA was used as the calibration gene of the internal reference. Use Primer Premier5.0 software to design the forward and reverse PCR primers respectively.
[0128] 1) ERα qPCR primer nucleic acid sequence (NCBI accession number: NM_000125)
[0129] Forward: 5'-ACCGAAGAGGAGGGAGAATG-3' (SEQ ID 1)
[0130] Reverse: 5'-AACAAGGCACTGACCATCTG-3' (SEQ ID 2)
[0131] 2) PR qPCR primer nucleic acid sequence (NCBI accession number: NM_000926)
[0132] Forward: 5'-TTCACCAGGTCAAGACATACAG-3' (SEQ ID 3)
[0133] Reverse: 5'-GTTGCCTCTCGCCTAGTTG-3' (SEQ ID 4)
[0134] 3) HER-2 qPCR primer nucleic acid sequence (NCBI accession number: NM_001005862)
[0135] Forward: 5'-TTACCAGTGCCAATATCCAGG-3' (SEQ ID 5)
[0136] Reverse: 5'-TCCAGAGTCTCAAACACTTGG-3' (SEQ ID 6)
[0137] 4) Nucleic acid sequence of internal reference gene 18S qPCR prime...
Embodiment 3
[0140] Example 3 qPCR amplification and establishment of detection standard curve
[0141] (1) Template preparation: The standard plasmid constructed in step 1 was serially diluted 10 times, and the dilution was used as a template. The specific dilution concentrations are: 1, 1 / 10, 1 / 100, 1 / 1000, 1 / 10000.
[0142] (2) qPCR reaction system
[0143] SYBR Green I 5 μL
[0144] Primer (Forward) 0.4μL
[0145] Primer (reverse) 0.4μL
[0146] Template 1 μL
[0147] Diluent 4.2uL
[0148] (3) Cyclic reaction:
[0149] Phase 1: Pre-denaturation
[0150] Repeat: 1 time
[0151] 95°C for 30 seconds
[0152] Stage 2: PCR reaction
[0153] Repetition: 40 cycles
[0154] 95℃ for 5 seconds
[0155] 60°C 30 seconds
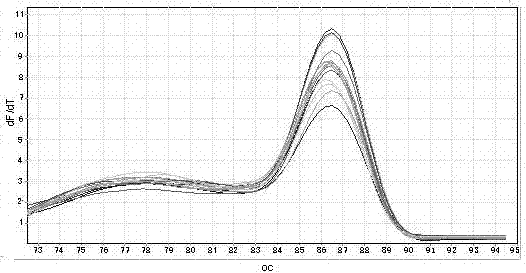
[0156] Phase: Dissolving
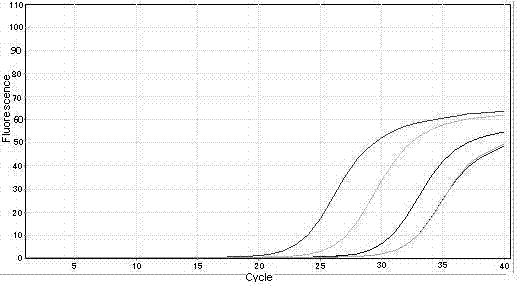
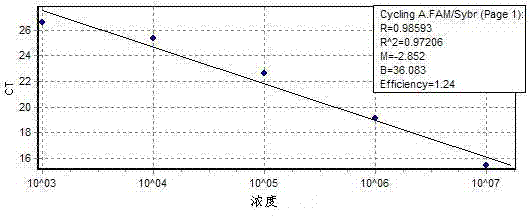
[0157] (4) Establish a standard curve: Fluorescence PCR collects fluorescent signals, and the standard curve is automatically generated by computer software, as shown in Figure 1(c), Figure 2(c), Figure 3(c), and Figure 4(...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com