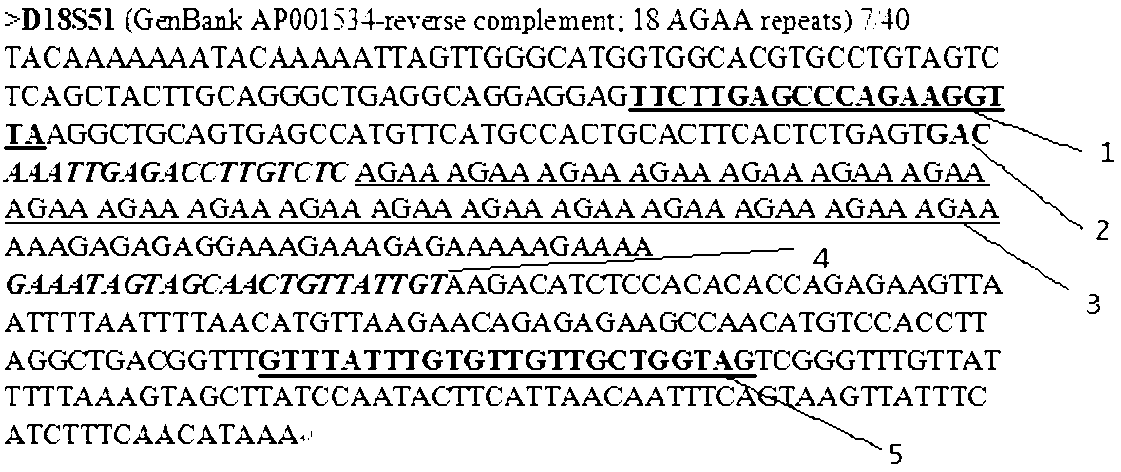STR (short tandem repeat) sequence high-throughput detection method with base selective controllable extension and detection reagent thereof
A detection method and selective technology, which is applied in the field of high-throughput DNA sequencing in biotechnology, can solve the problems of small throughput and inability to meet simultaneous detection, and achieve the goal of reducing complexity, reducing the number of base sites, reducing costs and consumption the effect of time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Using a high-throughput detection method for STR sequences with base-selective and controlled extension to perform multi-sample high-throughput gender detection on the STR sequence of the Amelogenin locus:
[0039] Using a high-throughput detection method for STR sequences with base-selective and controllable extension, the detection steps are:
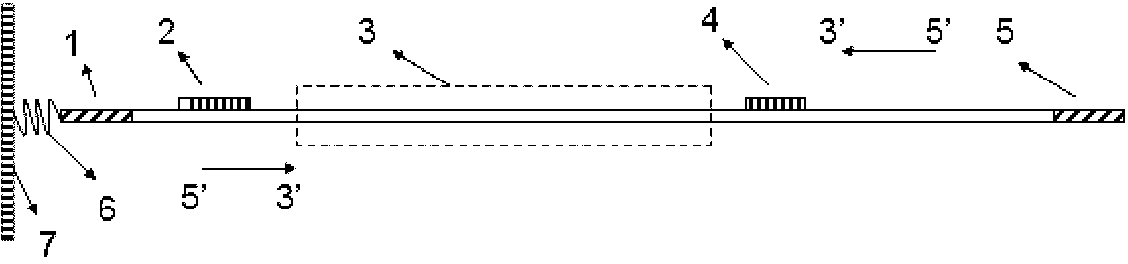
[0040] (1) Selection and amplification of the locus: Add a pair of PCR amplification primers for the Amelogenin locus together with the DNA template of the sample to be tested into the amplification system, control appropriate reaction conditions, and perform PCR reaction to obtain the Amelogenin gene The STR sequence amplification product of the locus. Wherein, the 5' end of one of the pair of amplification primers is modified with a connectable group. Different samples need to be amplified in independent amplification systems.
[0041](2) Preparation of the STR sequence library on the detection chip: by spottin...
Embodiment 2
[0050] Example 2: D3S1358, TH01, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, TPOX, SE33, D8S1179, D21S11, vWA using a high-throughput detection method of base-selective and controlled extension of STR sequences , STR sequences of 16 loci of FGA are used for identification and detection of sample separation, and the library preparation adopts the chip direct preparation method:
[0051] Using a high-throughput detection method for STR sequences with base-selective and controllable extension, the detection steps are:
[0052] (1) Selection and amplification of loci: add a pair of PCR amplification primers corresponding to the above 16 loci to the amplification system together with the DNA template of the sample to be tested, control appropriate reaction conditions, and perform PCR reaction. The STR sequence amplification product of the locus was obtained. Different samples need to be amplified in independent amplification systems.
[0053] (2) Prepara...
Embodiment 3
[0064] Example 3: F13A1, F13B, FES / FPS, HPRTB, LPL, D22S1045, DYS19, DYS385 a / b, DYS388, DYS389 I / II, DYS390, DYS391 by a high-throughput detection method of base-selective and controlled extension of STR sequences 17 loci, DYS392, DYS393, DYS434, DYS437, and Y-GATA-H4, were tested for mixed samples at the same time, and the library was prepared using the magnetic bead preparation method:
[0065] Using a high-throughput detection method for STR sequences with base-selective and controllable extension, the detection steps are:
[0066] (1) Selection and amplification of loci: Due to the mixed sample system, nucleic acid identification fragments need to be introduced to distinguish different samples and different loci. Firstly, the corresponding nucleic acid identification fragments are introduced into a pair of PCR amplification primers corresponding to the above-mentioned 17 loci, and then the amplification primers introduced with the nucleic acid identification fragments ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com