Method for eliminating dependence of methanol-induced promoter on single methanol carbon source
一种启动子、诱导型的技术,应用在生物工程领域,能够解决工业化生产麻烦、难满足氧气的需求、冷却能力要求越高等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
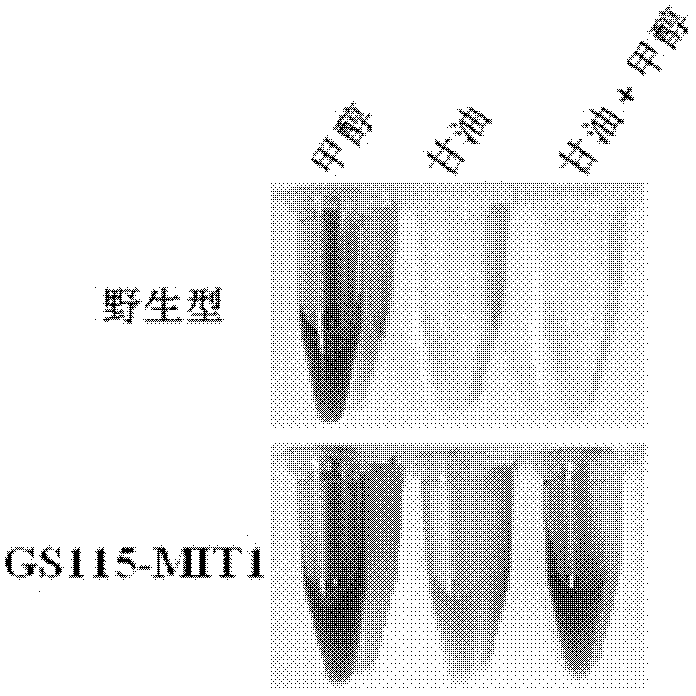
[0109] Example 1, Pichia pastoris strain GS115-MIT1 that glycerol can induce the expression of AOX1 promoter
[0110] 1. Construction of PpMIT1 overexpression plasmid
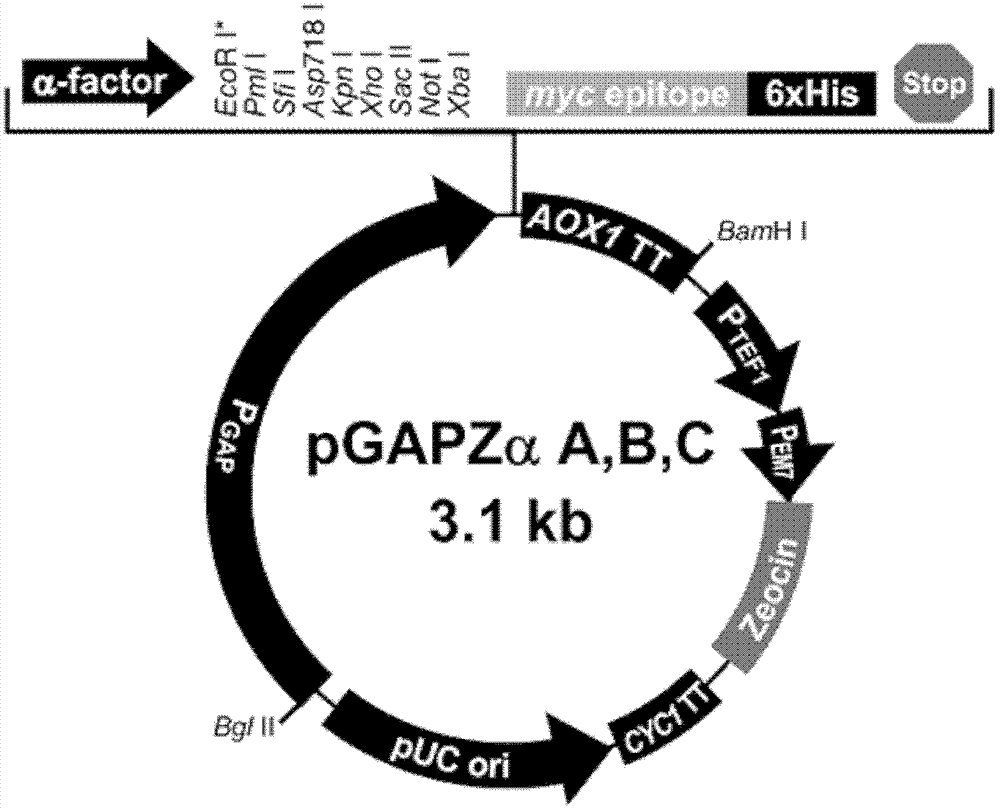
[0111] Using PCR, the method of restriction restriction ligation, with pGAPZαA ( figure 1 ) is a vector that inserts the PpMIT1 gene (SEQ ID NO: 1, the full length of the gene is 2667bp) at the Asu II / Sal I restriction sites downstream of the GAP promoter, and the resulting recombinant plasmid is called pGM plasmid.
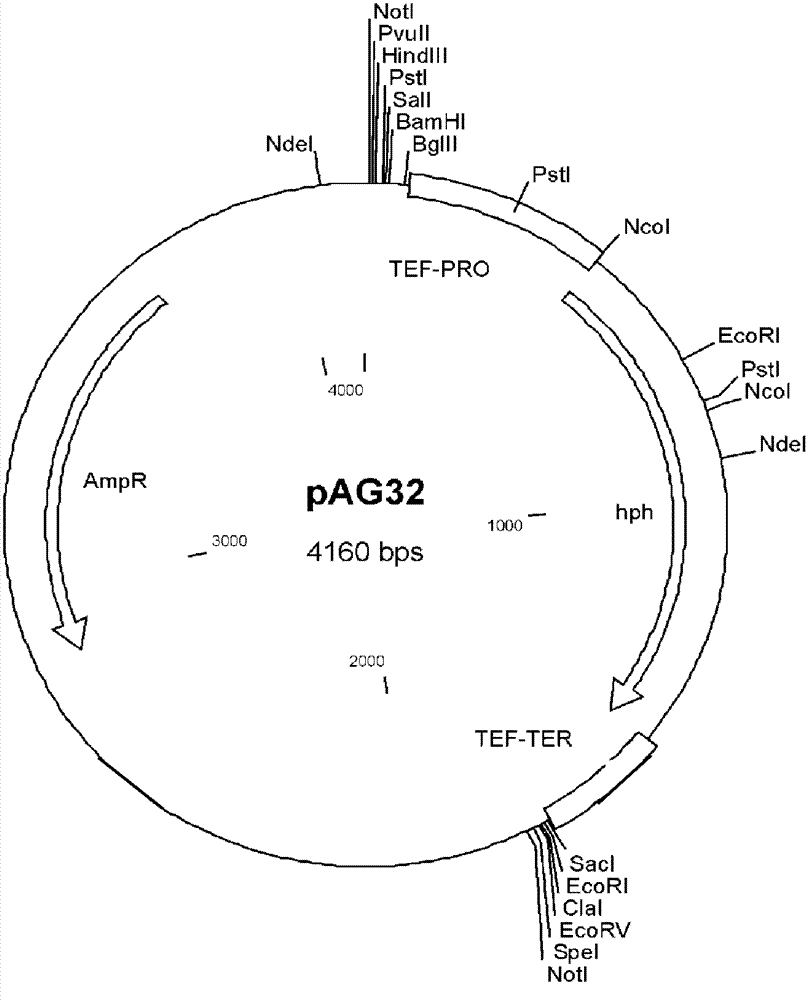
[0112] Using M1-GAP5 and M1-AOX1TT as primers (Table 1), the expression system of pGAP-PpMIT1-AOX1TT (full length 3615bp) was amplified from the pGM plasmid by PCR method, and pAG32( figure 2 ) is the vector inserted into the expression system at the Sac I / Spe I place. The obtained recombinant plasmid was pGMhph.
[0113] Table 1. Primer List
[0114]
[0115]
[0116] 2. Screening of electroporated Pichia pastoris and GS115-MIT1 strain
[0117] The PpMIT1 overexpression plasmid (pGMhp...
Embodiment 2
[0134] Example 2, Glycerol or Glucose Inducible AOX1 Promoter Expression Pichia strain
[0135] 1. Construction of HXS1 gene knockout plasmid
[0136] The Sh ble fragment of the Zeocin resistance gene was excised from the plasmid pGAPZαA with BamH I and Sal I enzymes, and converted to the pUC18 plasmid ( Figure 10 ) as a vector, the Zeocin resistance gene Sh ble fragment was inserted into the BamH I and Sal I enzyme-cut ligation, named pUC18-ble plasmid. Using the GS115 genome as a template, primers HS1-5F / HS1-5R were first used to amplify the 5' peripheral promoter region of the PpHXS1 gene (upstream of the start codon ATG), and the size of the amplified product was 728bp. Primers HS1-3F / HS1-3R were used to amplify the 3' end region of the PpHXS1 gene (downstream of the stop codon). The size of the amplified product was 1011 bp, and the fragments of the target size were recovered by cutting the gel separately.
[0137] The peripheral promoter DNA fragment at the 5' end of ...
Embodiment 5
[0155] Example 5. Pichia pastoris strain Δmig1ΔmigΔnrg1-MIT1 in which expression of AOX1 promoter can be induced by glycerol or glucose
[0156] 1. Construction of NRG1 gene knockout plasmid
[0157] Using the GS115 genome as a template, the primer 5'NRG1-F / 5'NRG1-R was used to amplify the 5' end peripheral region of the PpNIG1 gene to obtain a 320bp fragment, and the amplified product was named 5'NRG1, and the gel recovered 5'NRG1 fragment. The 5'NRG1 fragment obtained above was double-digested with Sac I and Sma I, and ligated with the pUC18 plasmid that was also double-digested with Sac I and Sma I to form pUC18 (SacI-5'NRG1-SmaI). The vector pUC18 (SacI-5'NRG1-SmaI) was transformed into competent Escherichia coli, and positive clones were screened by colony PCR.
[0158] Then, using primers HYG-F and HYG-R, the plasmid pRDM054 was used as a template to amplify the hygromycin B resistance gene HPH with a size of 1648bp. The HPH fragment obtained above was double digested...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com