Biosynthesis gene cluster of griseoviridin and viridogrisein and application of biosynthesis gene cluster
A technique for chlorophyll and griseogreen, which can be applied in the directions of plant genetic improvement, application, microorganism, etc., and can solve the problems of low yield and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] Extraction of Genomic DNA from Streptomyces griseoviridis NRRL 2427, which produces griseoviricin and viridomycin:
[0115] Inoculate the spores of fresh Streptomyces griseoviridis NRRL 2427 into 50 mL of TSB medium (tryptone 17g, plant peptone 3g, sodium chloride 5g, dipotassium hydrogen phosphate 2.5g, glucose 2.5g, add water to 1L, pH 7.2-7.4), 28-30°C, shake culture for about 2 days, centrifuge at 4000rpm for 10 minutes to collect mycelium. Wash the mycelia twice with STE solution (NaCl 75mM, EDTA 25mM, Tris-Cl 20mM), add 30mL of STE solution and lysozyme with a final concentration of 3mg / mL to the washed mycelium, vortex evenly, and store at 37°C Incubate for 3 hours, add proteinase K to a final concentration of 0.1-0.2mg / mL, mix well, incubate at 37°C for 10 minutes, add SDS to a final concentration of 1-2%, mix well, put in a 55°C water bath for about 1 hour, Invert several times during this period. Add an equal volume of phenol-chloroform-isoamyl alcohol (V / V / ...
Embodiment 2
[0117] Construction of Genomic Library of Streptomyces griseoviridis NRRL 2427, a Giseoviridin and Viridomycin Producer:
[0118] First, through a series of dilution experiments to determine the amount of restriction endonuclease Sau3A I, in a 20 μL system, containing 17 μL of Streptomyces griseoviridis NRRL 2427 genomic DNA, 2 μL of 10 × reaction buffer and 1 μL of different dilutions of For Sau3A I, the stop reaction was 4 μL of 0.5 mol / L EDTA and an appropriate loading buffer. Through exploration, it is determined that the enzyme activity unit of 0.025-0.05U is more appropriate. On this basis, a genomic DNA fragment slightly larger than 40kb was obtained by a large number of partial enzyme digestions, and dephosphorylated with a dephosphorylase.
[0119] The vector SuperCos l plasmid used to construct the library was first cut from the middle of the two cos sequences with restriction endonuclease Xba I, then dephosphorylated, and then used restriction endonuclease Bam from...
Embodiment 3
[0122] Positive clones containing biological genes synthesized by griseoviricin and viridomycin were screened from the genome library of Streptomyces griseoviridis NRRL 2427, a producer of griseoviricin and viridomycin:
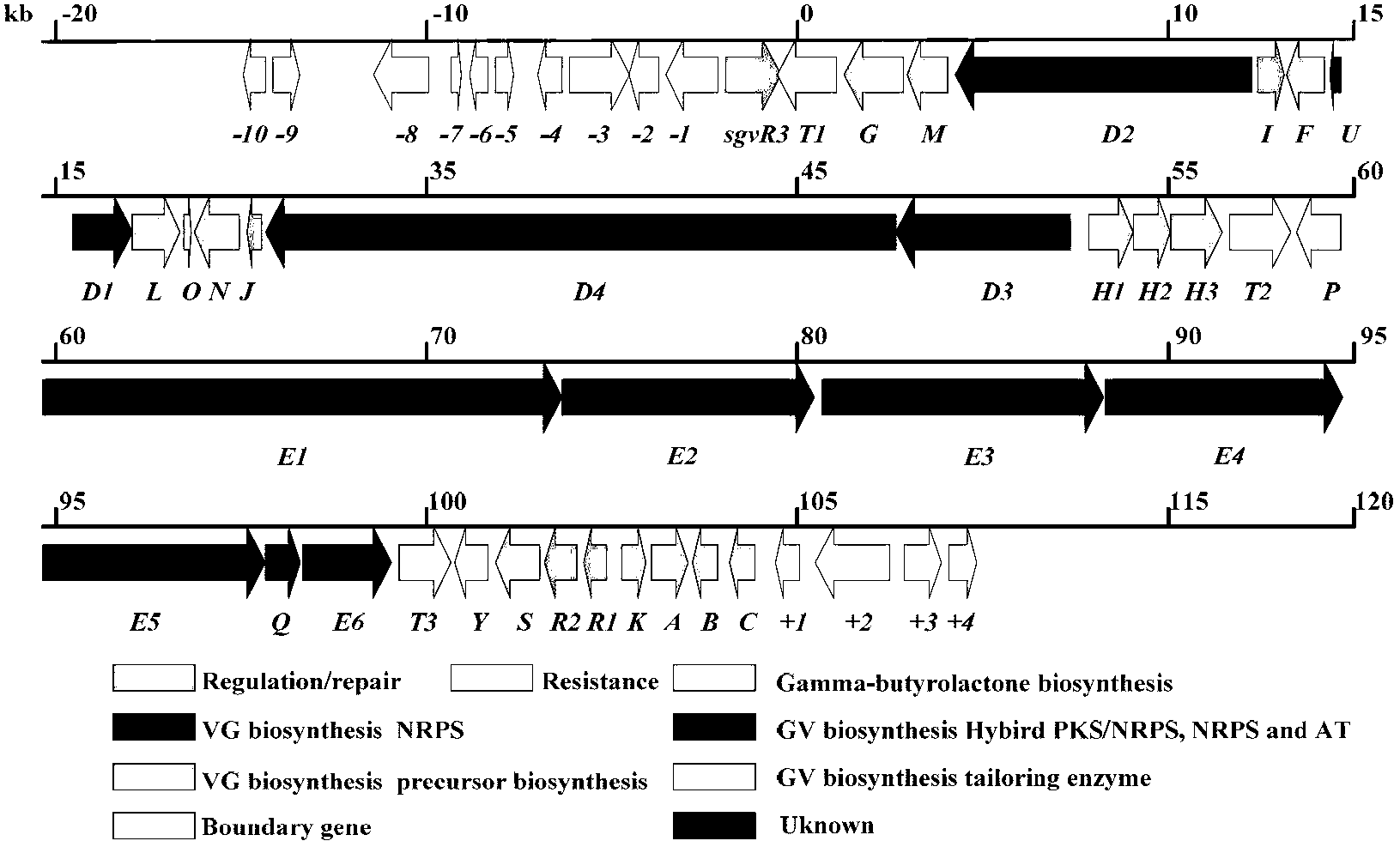
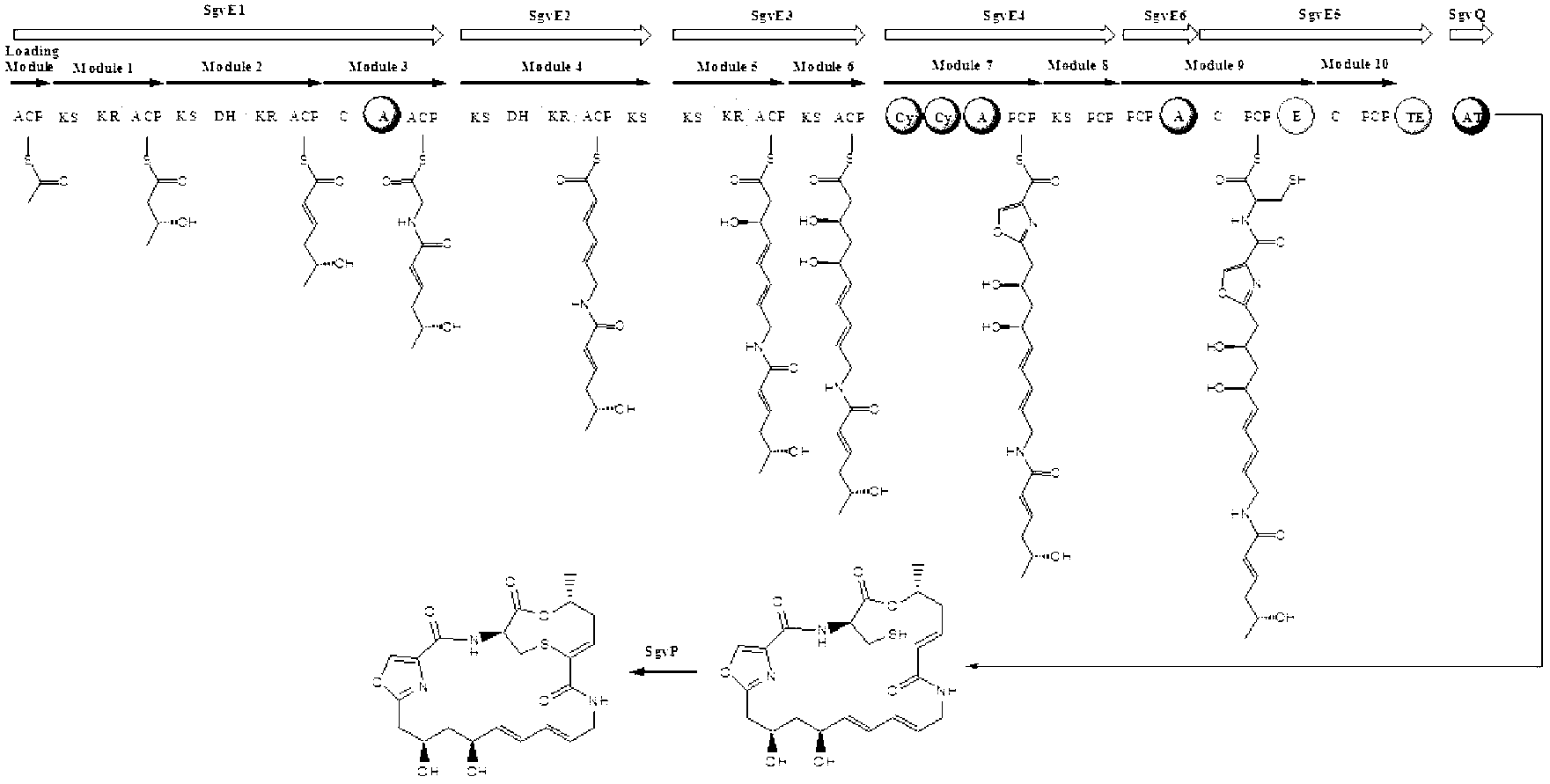
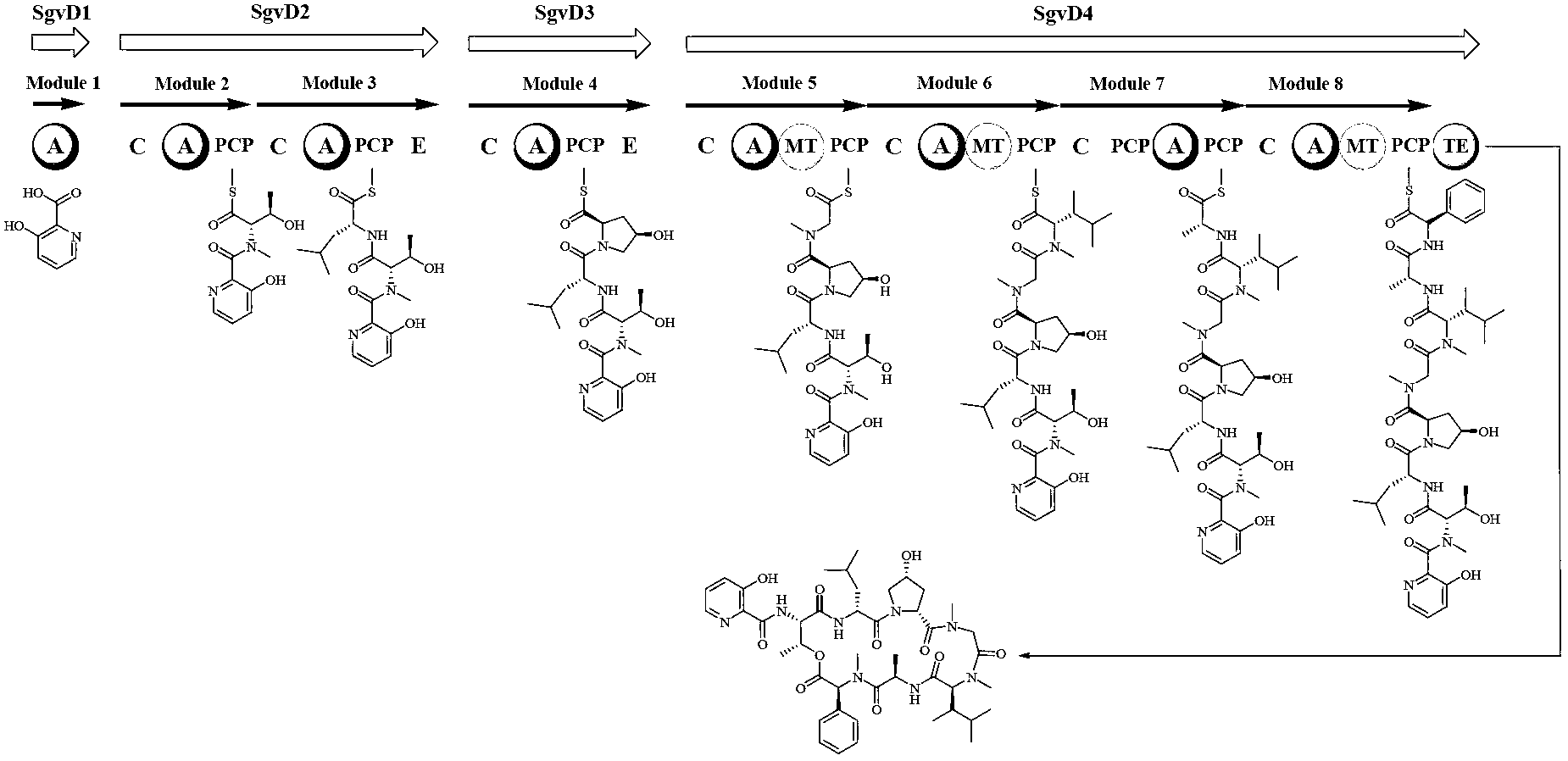
[0123] Genomic DNA of Streptomyces griseoviridis NRRL 2427 was sent to Macrogen Corporation of South Korea for genome-wide scanning and annotation. Based on the results of scanning and annotation, bioinformatics analysis was used to preliminarily determine the specific genes of griseoviridin and pyloridemicin biosynthetic gene clusters. position, and designed 5 pairs of screening primers (Table 4), screened 2400 clones by PCR, obtained 16 positive clones, and determined the positive clones containing the biosynthetic gene clusters of griseoviricin and chlorogreymycin, And sequenced, its nucleotide sequence is shown in SEQ ID NO.1, and the nucleotide sequence of the biosynthetic gene cluster of griseocide and viridocycin is shown in sequence 13530~ of sequence ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com