Method for producing microbial transglutaminase by use of pro-transglutaminase
A technology of transglutaminase proenzyme and transglutaminase enzyme is applied in the field of using transglutaminase proenzyme to produce microbial transglutaminase, and can solve the problem of unstable production, cumbersome process and unsatisfactory effect. and other problems, to achieve the effect of simplifying the fermentation production process, avoiding the degradation of enzymes, and achieving a good purification effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
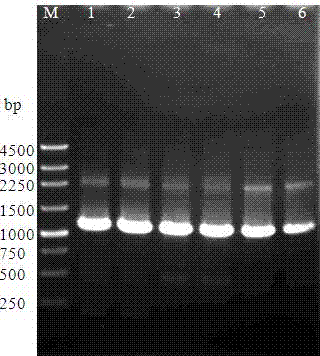
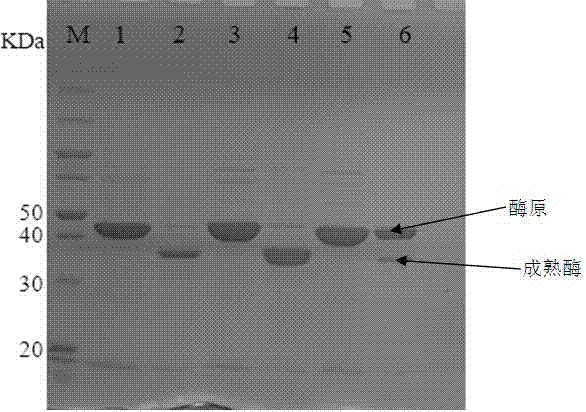
[0029] (1) Amplification of the transglutaminase progenase gene and introduction of the recognition sequence of bovine enterokinase: Streptomyces mobara ( Streptomyces mobaraensia ) The nucleotide sequence of the transglutaminase zymogen, with 1 mug Streptomyces mobara genomic DNA is the PCR reaction template, primers are SEQ ID No4, SEQ ID No5, SEQ ID No6, SEQ ID No7, the first five amino acids DSDDR of the mature peptide are changed to DDDDK, amplified and transgenic A DNA fragment corresponding to the size of the pro-aminoamidase gene, such as figure 1 As shown, M is Marker, 1 and 2 are the PCR products when the first five amino acids DSDDR of the mature peptide are changed to DDDDK in this embodiment, and the sequence of the transglutaminase zymogen gene fragment with the EK cleavage site See the sequence listing SEQ ID No1;
[0030] (2) Construct expression vector by gene cloning: pass the gene fragment with EK cleavage site obtained in step (1) through Nde I and ...
Embodiment 2
[0042] (1) Amplification of the transglutaminase zymogen gene and introduction of the recognition sequence of bovine enterokinase: obtained from NCBI Streptomyces mobara ( Streptomyces mobaraensia ) The nucleotide sequence of the transglutaminase zymogen, with 1 mu g Streptomyces mobara genomic DNA is used as the PCR reaction template, and the primers are SEQ ID No4, SEQ ID No8, SEQ ID No9, and SEQ ID No7, and DDDDK is directly inserted between the zymogen and the mature peptide to amplify and transglutaminase A DNA fragment matching the size of the zymogen gene, such as figure 1 As shown, M is Marker, 3 and 4 are the PCR products when DDDDK is directly inserted between the zymogen and the mature peptide in this example, and the sequence of the transglutaminase zymogen gene fragment with the EK cleavage site is shown in the sequence List SEQ ID No2;
[0043] (2) Construct expression vector by gene cloning: pass the gene fragment with EK cleavage site obtained in step (1) ...
Embodiment 3
[0049] (1) Amplification of the transglutaminase zymogen gene and introduction of the recognition sequence of bovine enterokinase: obtained from NCBI Streptomyces mobara ( Streptomyces mobaraensia ) The nucleotide sequence of the transglutaminase zymogen, with 1 mu g Streptomyces mobara genomic DNA is the PCR reaction template, primers are SEQ ID No4, SEQ ID No10, SEQ ID No11, SEQ ID No7, the last five amino acids of the zymogen, SFRAP, are changed to DDDDK to amplify and transglutaminase A DNA fragment matching the size of the zymogen gene, such as figure 1 As shown, M is Marker, and 5 and 6 are the PCR products when the last five amino acids of the zymogen were changed from SFRAP to DDDDK in this embodiment. List SEQ ID No3;
[0050] (2) Construct expression vector by gene cloning: pass the gene fragment with EK cleavage site obtained in step (1) through Nde I and xho I double digested, cloned into the prokaryotic expression vector pET22b(+), constructed the expressi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com