Pestalotia funereal pathogenic bacteria molecule detection kit and use method thereof
A detection kit and molecular detection technology, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of long cycle and achieve the effect of simple and fast operation, strong specificity, and improved efficiency and scope
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: the detection of pine red blight in plant tissue
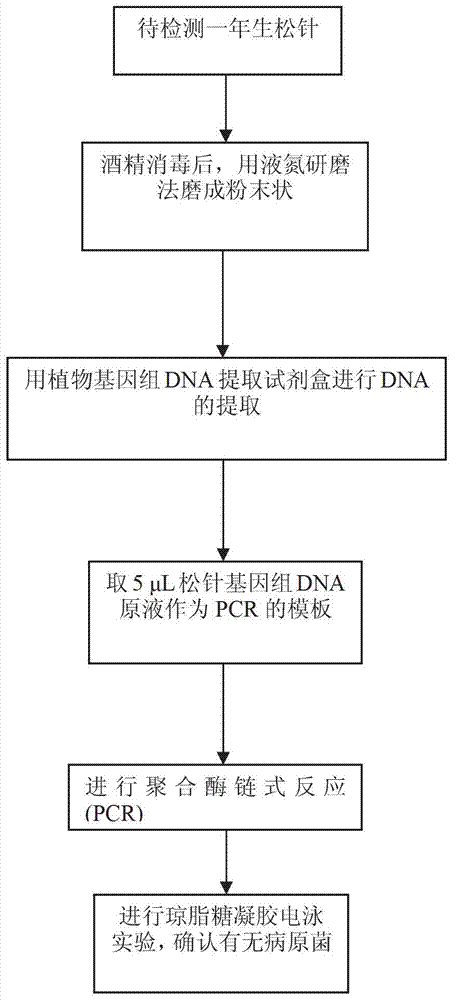
[0032] (1) After surface disinfection, the seemingly healthy samples were cut into three parts according to the leaf tip, leaf center and leaf base, and transferred to a sterilized mortar respectively, and were extracted by liquid nitrogen grinding method combined with a plant genomic DNA extraction kit. Extraction, the total DNA of the sample is used as a template for the next step of PCR;
[0033] (2) According to the design of the ITS region sequence (AF405299.1) of the bacteria already available in Genbank, a pair of PCR primers (AF-F / R) with high specificity were screened out, respectively:
[0034] Upstream primer F (5'-3'): GTCAACCAGCGGAGGGAT 18bp
[0035] Downstream primer R (5'-3'): CGCCGTTGTATTTCAGGAG 19bp
[0036] (3) 50 μL of PCR reaction system includes: 5 μL pine needle genomic DNA stock solution, 25 μL Premix, 2 μL of each primer (10 μmol / L), ddH 2 O 16 μL. The reaction was carried out o...
Embodiment 2
[0038] Embodiment 2: the specificity amplification of primer to pine wilt
[0039] 50 μL of PCR reaction system includes: 5 μL pine needle genomic DNA stock solution, 25 μL Premix, 2 μL of each primer (10 μmol / L), ddH 2 O 16 μL. The reaction was carried out on an iCycler-PCR instrument, and the program was: denaturation at 95°C for 4 minutes; denaturation at 95°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min, and 35 cycles; extension at 72°C for 7 min, and storage at 10°C.
[0040] Test results
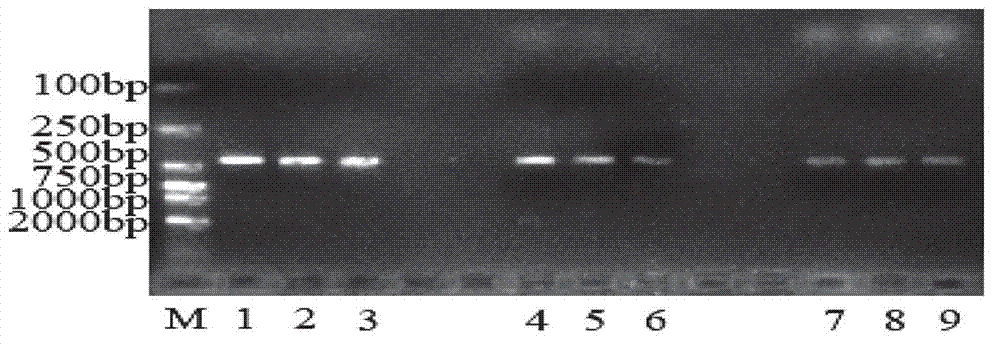
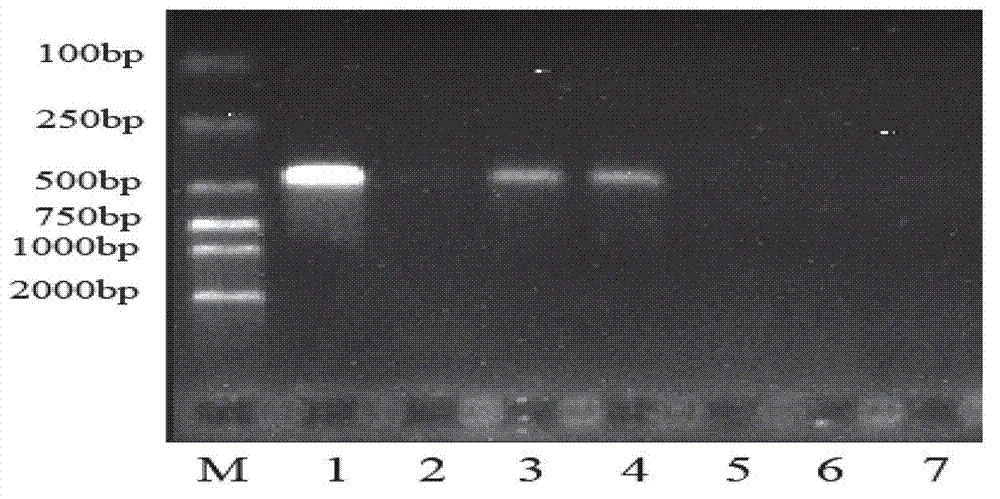
[0041] The specificity of detection: Use primer AF-F / R to carry out PCR reaction simultaneously to pine red blight and the arboreal fungus hyphae DNA that has been reported in the laboratory (reaction results see figure 2 ), the amplified band of about 420bp, while for Pestalotiapsis lespedezae, Pestalotiapsis lawsoniae, Lophodermiμm conigenμm, and loose needles The amplification results of Lophodermi μm pinastri and Fusari μmoxyspor μm showed darker or no band...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com