Plant in-vitro ubiquitin protein degradation system and application thereof
A technology of ubiquitin protein and protein, which is applied in the field of plant in vitro ubiquitin protein degradation system, which can solve the problems of single, false negative, and inability to detect E3 protein activity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
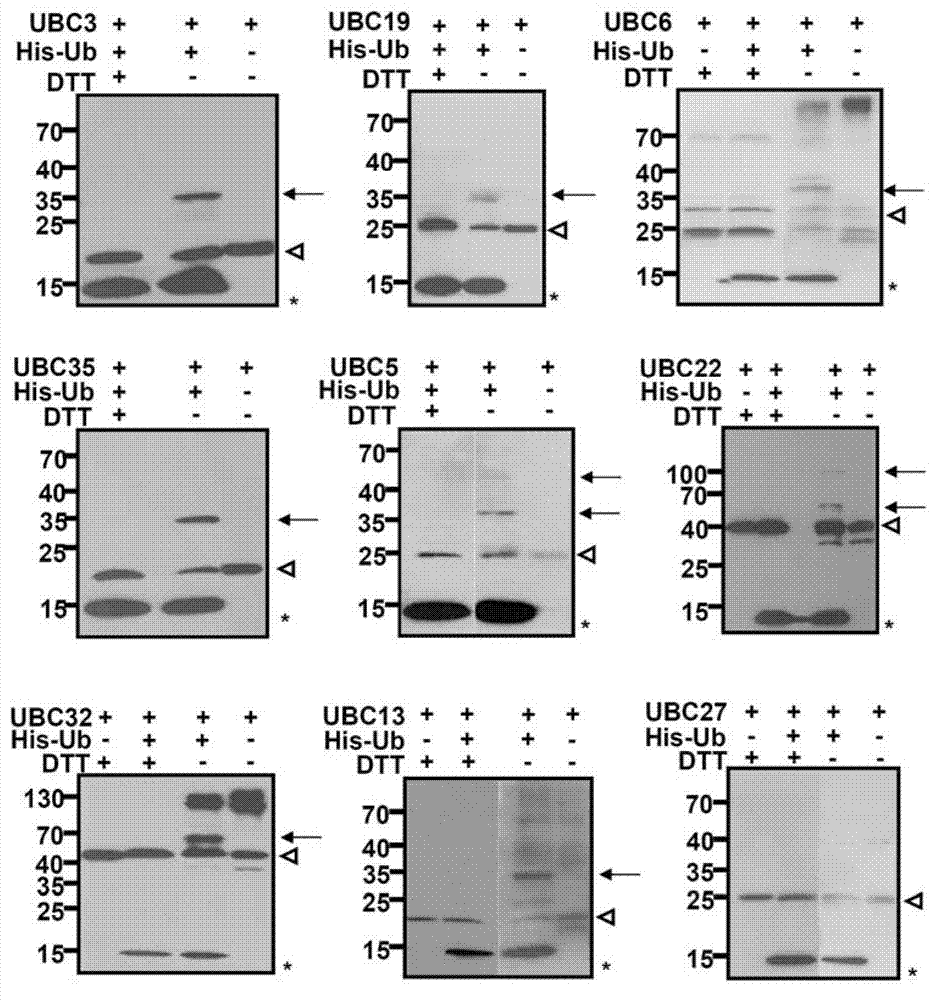
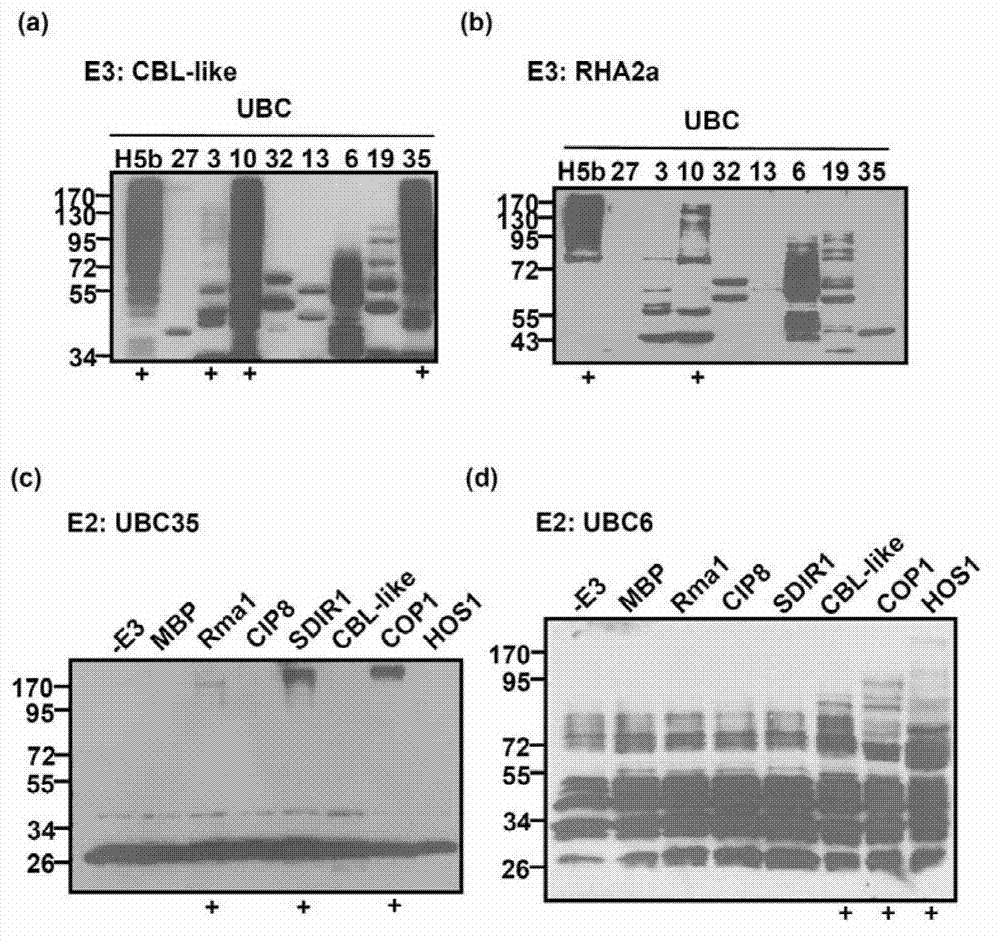
Examples
Embodiment 1
[0089] Embodiment 1, the construction of recombinant expression vector
[0090] 1. Preparation of recombinant plasmid pET28a-UBA2
[0091] 1. Design primers for amplifying the full CDS sequence of Arabidopsis thaliana ubiquitin activating enzyme UBA2 gene (GenBank: U40566.1, nucleotide sequence from 414 to 3647 at the 5' end, sequence 2) and separate Add EcoRI and SalI restriction sites.
[0092] 2. Use the primers in step 1 to clone the full CDS sequence of the UBA2 gene by PCR, and digest it with restriction enzymes EcoRI and SalI.
[0093] 3. Digest the pET28a vector with restriction endonucleases EcoRI and SalI to obtain the vector backbone.
[0094] 4. Ligate the digested product of step 2 with the vector backbone of step 3 to obtain the recombinant plasmid pET28a-UBA2. Sequencing showed that the recombinant plasmid pET28a-UBA2 was inserted into GenBank: U40566.1 between the EcoRI and SalI sites of the backbone vector pET28a, from nucleotide sequence 414 to 3647 at the...
Embodiment 2
[0149] Embodiment 2, the preparation of albumen
[0150] 1. Expression and purification of 6×His fusion protein and UBCH5b protein
[0151] 1. Prepare Escherichia coli Bl21(DE3) competent cells.
[0152] 2. The recombinant plasmids prepared in Step 1 and Step 6 of Example 1 and using pET28a as the vector backbone were transformed into Bl21(DE3) competent cells by heat shock method respectively.
[0153] 3. Pick 2-3 single clones and culture them overnight in 3-10 mL LB liquid medium supplemented with corresponding antibiotics at 37°C and 200 rpm.
[0154] 4. Add the overnight cell culture to 200-500mL LB liquid medium containing corresponding antibiotics and 0.2% (0.2g / 100mL) glucose, so that the OD600 is about 0.1.
[0155] 5. Shake culture at 18°C until OD600 is 0.5-0.6, add inducer isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.2mM; induce at 18°C for 12-16h.
[0156] 6. Centrifuge at 4000 rpm for 10 minutes at 4°C to collect bacteria.
[0...
Embodiment 3
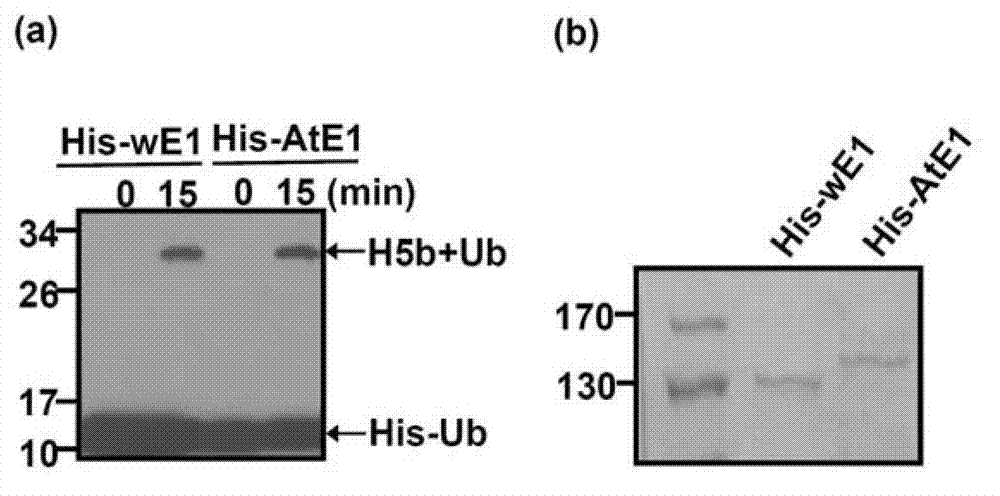
[0180] Activity detection of the Arabidopsis thaliana ubiquitin activating enzyme and the Arabidopsis ubiquitin protein purified in Example 3 and Example 2
[0181] In this example, the activities of the Arabidopsis thaliana ubiquitin activating enzyme and the Arabidopsis ubiquitin protein purified in Example 2 were detected according to the following method:
[0182] (1) Take two 1.5mL Eppendorf tubes and record them as sample 1 and sample 2 respectively. Add 0.2 μg of human ubiquitin-conjugating enzyme (UBCH5b) obtained in Example 2, 3-5 μg of Arabidopsis wild-type monomeric ubiquitin protein (His-Ub) and 1 μL of 20× reaction buffer ( Formulation: 1M Tris pH7.5, 100mM ATP, 50mM MgCl 2 , 2mM DTT, the solvent is water). In addition, 0.05 μg of Arabidopsis thaliana ubiquitin activating enzyme UBA2 (His-AtE1) was added to sample 1, and the same amount of wheat ubiquitin activating enzyme (His-wE1) was added to sample 2. Add ddH to both samples 2 O to a total volume of 20 μL....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com