Sickle-alfalfa chloroplast cold-response protein (MfcpCOR14) and coding gene and application thereof
A yellow alfalfa, chloroplast technology, applied in application, genetic engineering, plant genetic improvement and other directions, can solve the problem of little research on isolation of cold resistance genes, and achieve the effect of improving cold resistance and strong light resistance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Cloning of embodiment 1MfcpCOR14
[0051] 1. Preparation of Medicago cDNA template
[0052] Seeds of Medicago falcata were soaked in hot water at 60°C for 5 minutes, and germinated in the dark at 28°C. When the seeds were white, they were sown in plastic pots (15 cm in diameter) filled with soil and cultivated in the greenhouse. Light, temperature 25-28 ℃. Put the 8-10-week-old Medicago sativa plants into an artificial climate box at a temperature of 5°C (light intensity of 200 μmol·m -2 the s -1 ), the light / dark cycle was 12 hours of light, and the low temperature treatment was carried out at 5°C for 2 consecutive days. Mature leaves of Medicago sativa were taken, and total RNA was extracted by TRE-Trizol method, reverse-transcribed with M-MLV reverse transcriptase kit (Promega Company) to obtain reverse-transcribed first-strand cDNA as a template, and stored at -20°C for future use.
[0053] 2. Design and amplify the primers of MfcpCOR14 gene
[0054] According ...
Embodiment 2
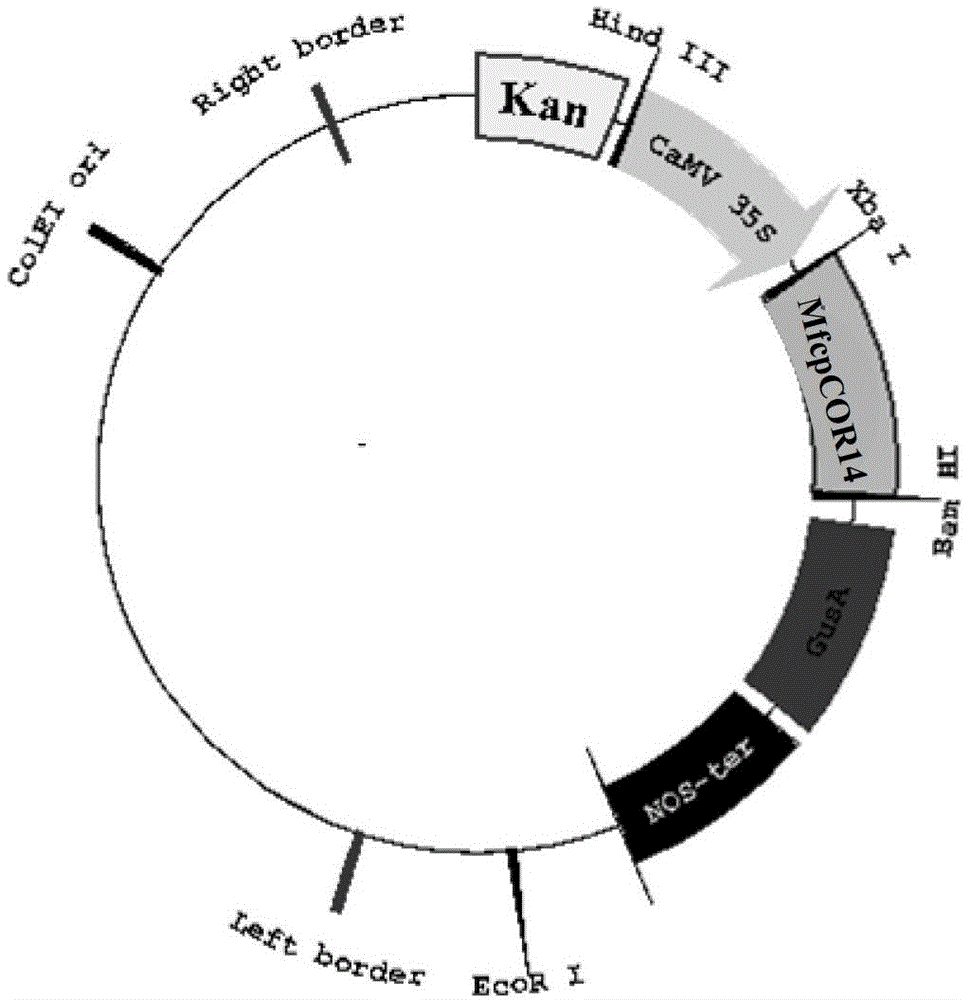
[0067] Example 2 Construction of the plant expression vector (pBI-MfcpCOR14) of the MfcpCOR14 gene
[0068] Since the MfcpCOR14 gene is reversely connected to the pMD20-T vector, design amplification primers with BamHI and XbaI restriction sites according to the flanking sequences of the insertion site of the pMD20-T vector:
[0069] The upstream primer pMD1 introducing the BamHI restriction site: GATC GGATCC GAGGATCTACTAGTCATATGG;
[0070] Mutate the original BamHI restriction site and introduce the downstream primer pMD2 of the XbaⅠ restriction site: GTGA TCTAGA GCTCGGTACCCGG AAA ATCCGA;
[0071] The cloning vector pMD-MfcpCOR14 constructed in Example 1 was used as a template, and pMD1 and pMD2 were used as upstream and downstream primers to perform PCR amplification of the MfcpCOR14 gene, and the amplified MfcpCOR14 gene fragment was purified and recovered.
[0072] The MfcpCOR14 gene fragment and the pBI121 expression vector recovered above were double-digested with ...
Embodiment 3
[0073] Example 3 MfcpCOR14 expression induced by low temperature
[0074] 1. Obtaining the Medicago sativa template under low temperature conditions
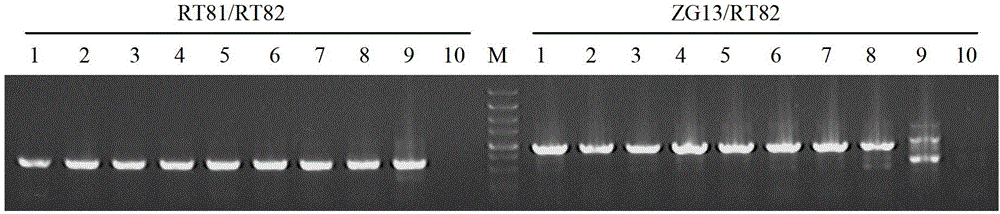
[0075] Put the 8-10-week-old Medicago sativa plants into an artificial climate chamber at a temperature of 5°C (200 μmol·m -2 the s -1 ), the light and dark time were both set to 12 hours of light. Continuous treatment for 4 days, low temperature treatment at 5°C. Take the mature leaves of low temperature treatment for 0h, 12h, 24h, 48h, and 96h respectively, add liquid nitrogen to grind the samples, extract the total RNA of the leaves with TRE-Trizol reagent, and use PrimeScript TM RT reagent Kit (TaKaRa Company) was reverse-transcribed to obtain reverse-transcribed first-strand cDNA as a cDNA template, which was stored at -20°C for future use.
[0076] 2. Design specific detection primers
[0077] According to the cDNA sequence of MfcpCOR14 gene and the sequence of truncated alfalfa Actin (MTR3g095530), quantitative prime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com