Alkylation derivatization reagent and application of alkylation derivatization reagent in peptide fragment marking and mass spectrometric detection
A derivative reagent and mass spectrometry detection technology, which is applied in the field of biomacromolecule labeling, can solve the problems of limited improvement in the detection sensitivity of peptide mass spectrometry, and achieve the effects of low cost, increased charge number, and simple synthesis process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
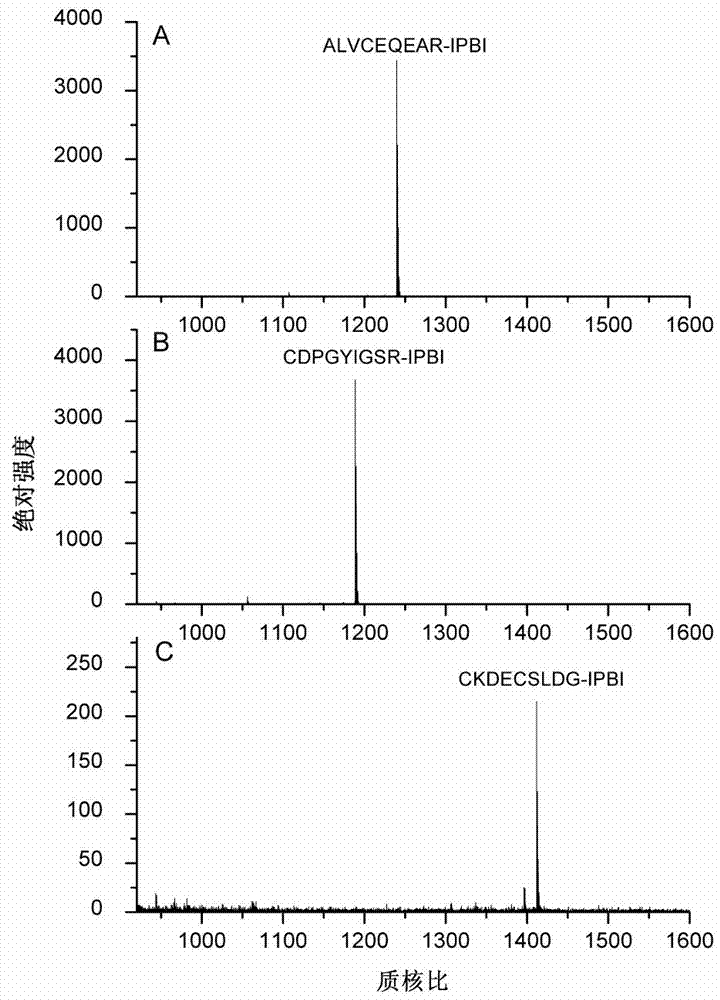
[0043] Tris-HCl buffer solution with a concentration of 50mmol / L and a pH of 8.4 was added to the peptide ALVCEQEAR to prepare a peptide solution with a concentration of 0.33mg / mL. Take 15 μL of the peptide solution, add 10 μL of tris(2-carboxyethyl)phosphine hydrochloride solution with a concentration of 20 mmol / L, and react at 50° C. for 1 hour. After cooling to room temperature, 50 μL of IPBI with a concentration of 50 mmol / L was added to the above solution, and the reaction was continued at 37° C. for 2 hours. After the reaction was completed, samples were directly sampled for MALDI-TOF MS analysis. The results showed that the derivation efficiency of the peptide ALVCEQEAR was 100% ( figure 1 Middle A panel).
Embodiment 2
[0045] Add 50 mmol / L Tris-HCl buffer solution with a pH of 8.4 to the peptide CDPGYIGSR to prepare a 0.33 mg / mL peptide solution. Take 15 μL of the peptide solution, add 10 μL of tris(2-carboxyethyl)phosphine hydrochloride with a concentration of 20 mmol / L, and react at 50° C. for 1 hour. After cooling to room temperature, 50 μL of IPBI with a concentration of 50 mmol / L was added to the above solution, and the reaction was continued at 37° C. for 2 hours. After the reaction was completed, samples were directly sampled for MALDI-TOF MS analysis. The results showed that the derivatization efficiency of the peptide CDPGYIGSR was 99.4% ( figure 1 Middle B panel).
Embodiment 3
[0047] Tris-HCl buffer solution with a concentration of 50mmol / L and a pH of 8.4 was added to the peptide CKDECSLDG to prepare a 0.33 mg / mL peptide solution. Take 15 μL of the peptide solution, add 10 μL of tris(2-carboxyethyl)phosphine hydrochloride solution with a concentration of 20 mmol / L, and react at 50° C. for 1 hour. After cooling to room temperature, 50 μL of IPBI with a concentration of 50 mmol / L was added to the above solution, and the reaction was continued at 37° C. for 2 hours. After the reaction was completed, samples were directly sampled for MALDI-TOF MS analysis. The results showed that the derivatization efficiency of the peptide CKDECSLDG was 100% ( figure 1 Middle C panel).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com