Method of identifying protein phosphorylation modification sites on the basis of tandem mass spectrometry
A tandem mass spectrometry and phosphorylation technology, which is applied in the field of identification of protein phosphorylation modification sites based on tandem mass spectrometry, can solve the problem of high limitations, and achieve the effect of accurate identification and improved reliability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
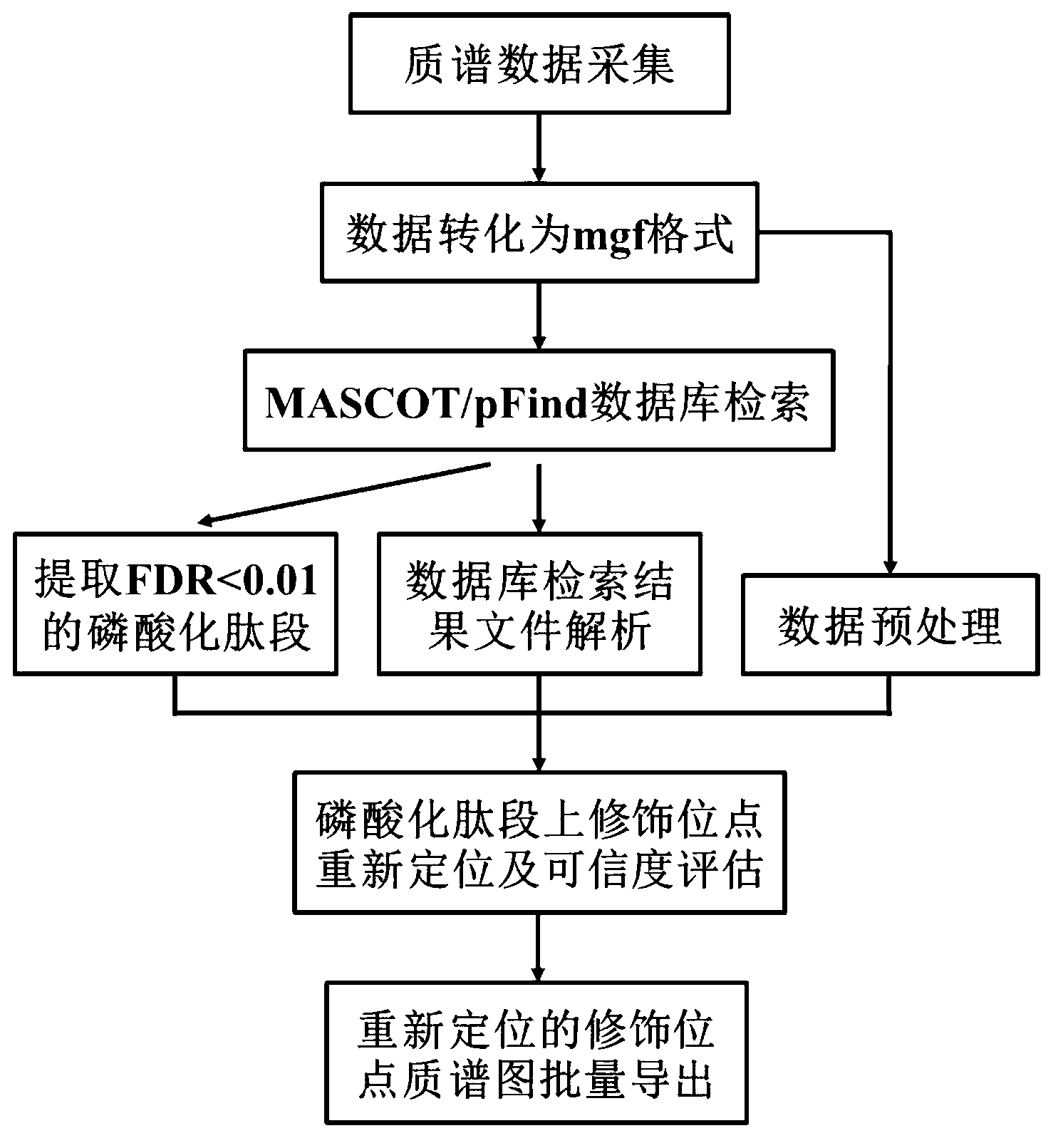
[0030] Example 1: Identification of protein phosphorylation modification sites based on Bruker ion trap mass spectrometry (amazon) data, the steps are as follows:
[0031] 1) Phosphopeptide database search:
[0032] The present invention is based on MASCOT and pFind database retrieval results, therefore, it is necessary to perform database retrieval first. The mass spectrometry data used in this example comes from the ion trap mass spectrometer amazon ETD of Bruker Company. The original data collected by the mass spectrometer is in the format of "*.yep". Use the company's software DataAnalysis4.0 software to perform peak conversion, export the results and save them as mgf format files, and then use the open source free software ProteoWizard to convert the data into standard mgf format file.
[0033] Open the local database search software MASCOT or pFind, import the mgf format data file, and set the relevant search parameters: trypsin digestion (trypsin), cysteine alkylati...
Embodiment 2
[0052] Example 2: Identification of protein phosphorylation modification sites based on AB Sciex company TripleTOF4600 mass spectrometry data
[0053] In this embodiment, the data format collected by TripleTOF mass spectrometry is ".wiff", and the open source free software ProteoWizard can be directly used to convert the data into a standard mgf format file. Then search the MASCOT and pFind databases, relocate the phosphorylation modification sites, evaluate the reliability, and automatically export the spectra in batches. The specific implementation method is the same as that in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com