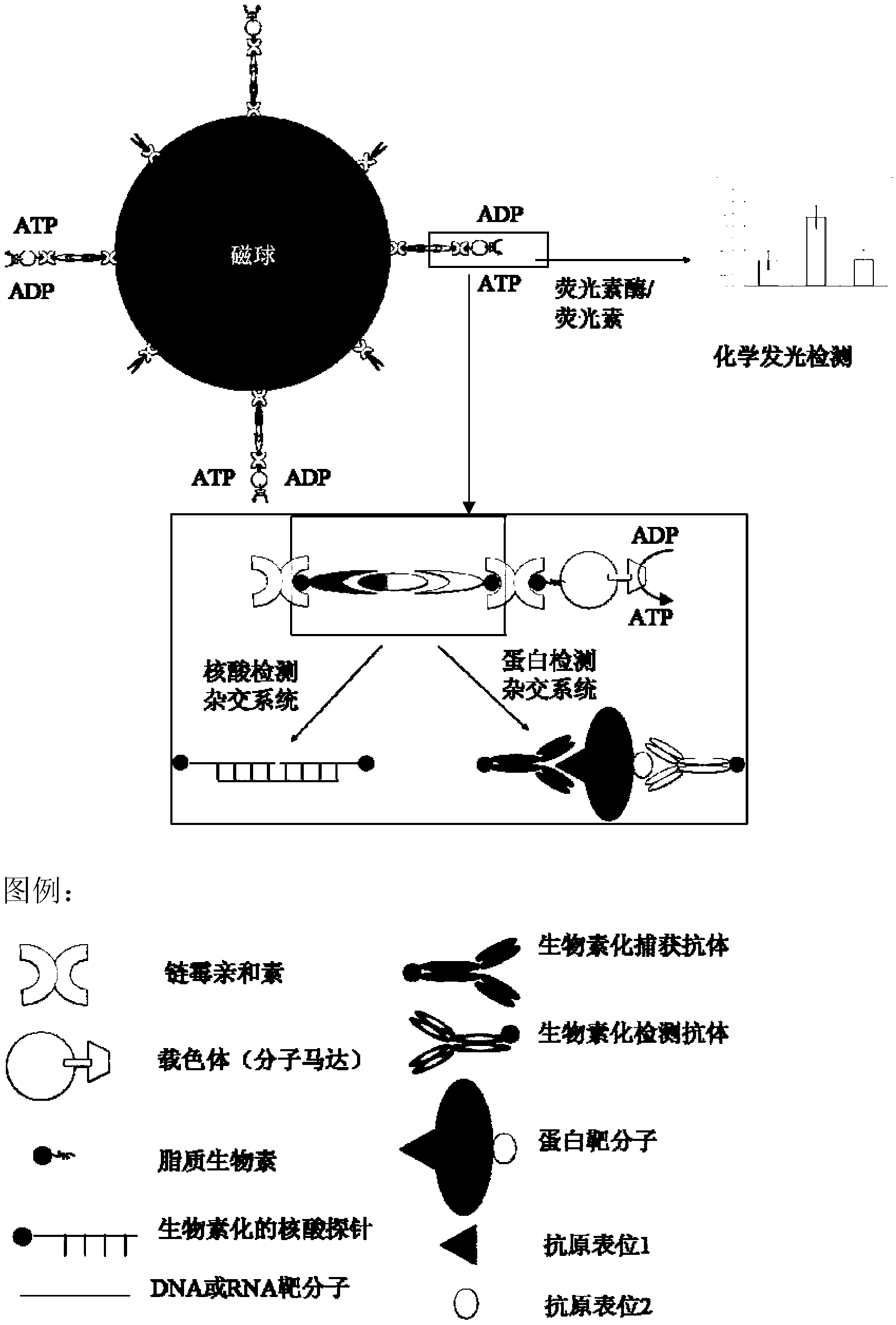
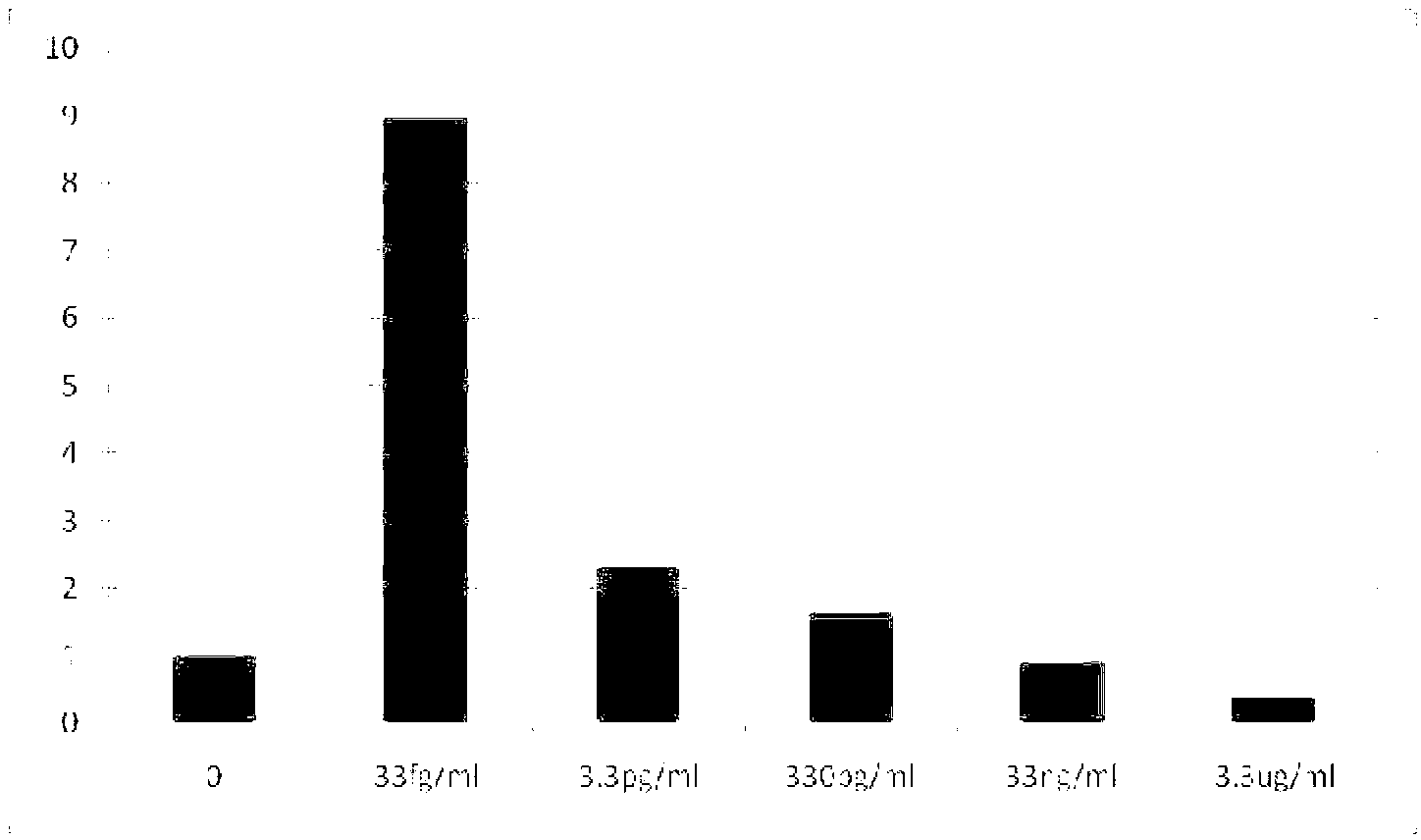Biosensor construction and detection method based on molecular motor, magnetic enrichment and double-probe hybridization
A molecular motor, biotin technology, applied in biochemical equipment and methods, biological testing, microbial determination/inspection, etc., can solve problems such as inability to detect multiple times, unfavorable comparisons, and limited detection sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, the general method of detection biomacromolecule
[0039]1. Preparation of ATP molecular motor:
[0040] Thermomicrobium roseum strains (ATCC27502, purchased from ATCC Species Bank, USA) were inoculated into liquid medium (see Table 1) at a ratio of 1:100, and cultured at 60° C. and 150 rpm for 24 hours. Then 4000rpm, 30min, 4 ℃ centrifugation to collect bacteria. With extraction buffer (20mM Tris-ClpH8.0, 100mM NaCl, 2mM MgCl 2 , 1 mM DTT) to resuspend the cells, and centrifuge to remove the supernatant (6000 rpm, 10 min, 4°C). Add extraction buffer to resuspend the cells (approximately 1 g of cells was added to 10 ml of buffer), then add PMSF at a final concentration of 1 mM, and sonicate for 30 min on ice (sonication for 5 s, stop for 8 s, power 300 W). Centrifuge the broken cells (25,000g, 30min, 4°C), remove the precipitate and get the supernatant. The supernatant was ultracentrifuged (145,000g, 1h, 4°C), and the precipitate was obtained as a chro...
Embodiment 2
[0075] Example 2, taking C-reactive protein (CRP) as an example to detect protein molecules
[0076] 1. Prepare the chromophore according to the method described in Example 1.
[0077] 2. Preparation of capture antibody and detection antibody: the C-reactive protein (CRP) used was purchased from meridian lifescience (A97201H). The capture antibody used is CRP monoclonal antibody M86842M (meridianlifescience), and the detection antibody used is CRP monoclonal antibody M86284M (meridianlifescience). The biotinylation process is as follows: add 2μl (+)-BiotinN-hydroxysuccinimide ester (10mM) (sigma, product number H1759) to 200μl CRP capture antibody M86842M (5mg / ml) and CRP detection antibody M86284M (7.5mg / ml) respectively , incubated at room temperature for 4 hours, free (+)-Biotin N-hydroxysuccinimide ester was removed by PBS dialysis, and the dialysate (PBS) was changed three times, each dialysis was 8 hours. The dialyzed biotinylated antibody was collected and added with ...
Embodiment 3
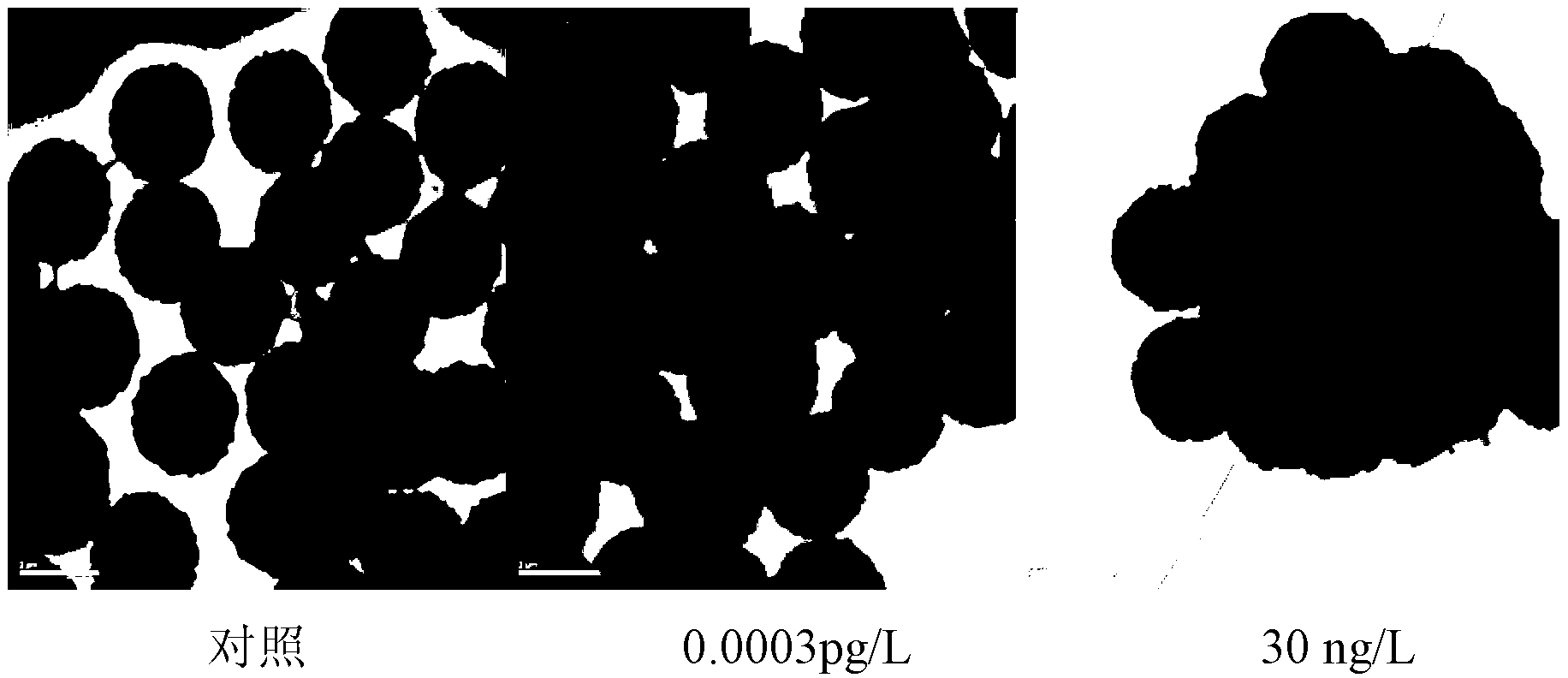
[0091] Example 3. Taking miR141 as an example to detect nucleic acids at different concentrations
[0092] A microRNA (miR141) is used as a target nucleic acid molecule for detection.
[0093] 1. Prepare the chromophore according to the method described in Example 1.
[0094] 2. Preparation of miR141 capture probe and detection probe:
[0095] miR141 (the sequence of miR141 is: UAACACUGUCUGGUAAAGAUGG) was synthesized as a standard sequence at Shanghai Jima Biological Co., Ltd., and diluted to 20uM with diethyl pyrocarbonate (0.1% in DEPC water) as a storage solution. Before the experiment, the storage solution was diluted with DEPC water Diluted into different concentrations (respectively 0, 1nM, 5nM, 10nM, 20nM, 40nM).
[0096] Synthesis of biotinylated capture probes and detection probes (Shanghai Gemma Biological Co., Ltd.):
[0097] Capture probe miRNA141-1: biotin-AAAAAAAAAA CCATTCTTTACC
[0098] Detection probe miRNA141-2: AGACAGTGTTA AAAAAAAAAAA-biotin
[0099]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com