Application of miR-17-5p and miR-20a in preparation of etoposide drug-resistant reversal agent
A mir-20a, Poside-sensitive technology, applied in anti-tumor drugs, drug combinations, pharmaceutical formulations, etc., can solve the problems of p53 deletion sites, mutations, severe bone marrow suppression, gastrointestinal reactions, liver and kidney damage, etc. Achieve the effect of huge application value, strong operability, and simple and easy technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
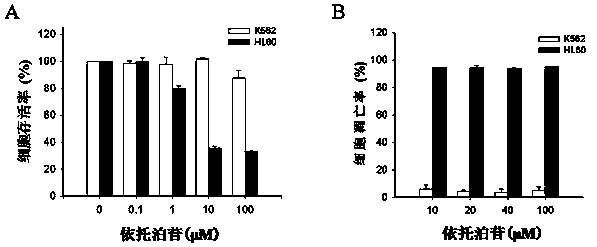
[0039] Example 1 Differences in sensitivity between K562 cells and HL60 cells to the chemotherapeutic drug etoposide
[0040] Divide K562 cells and HL60 cells into 3×10 5 / mL density was inoculated in 96-well plates, and after 24 hours of culture, 0.1-100 μmol / L etoposide was added to treat for 24 hours (with DMSO as the control), and then the cell viability was detected by MTT. See the experimental results figure 1 A, Etoposide could not effectively reduce the survival rate of K562 cells at each concentration tested, and 100 μmol / L etoposide could only reduce the survival rate of K562 cells by about 10%. On the contrary, the inhibitory effect of etoposide on HL60 cells was positively correlated with the dose of the drug, and 100 μmol / L etoposide could reduce the survival rate of HL60 cells to less than 40%.
[0041] Divide K562 cells and HL60 cells into 3×10 5 / mL density was inoculated in a 48-well plate, and after 24 hours of culture, 10-100 μmol / L etoposide was added ...
Embodiment 2
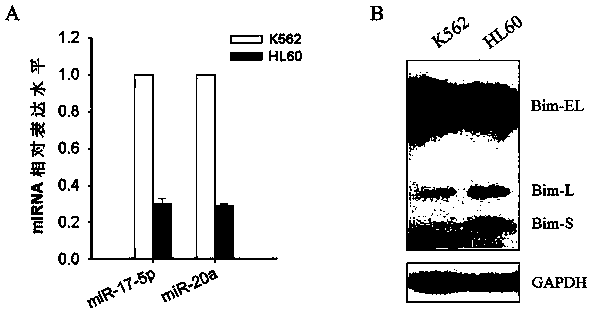
[0042] Example 2 Expression difference of miR-17-5p, miR-20a and Bim-S protein in K562 cells and HL60 cells
[0043] K562 cells and HL60 cells in logarithmic growth phase were collected, and total RNA was extracted using Trizol Reagent (Invitrogen, USA). The RNA concentration was measured by UV spectrophotometer. Then the extracted total RNA was reverse-transcribed (1 hr at 42 °C, 5 min at 95 °C) and real-time quantitative PCR reaction with miRCURY LNA® Universal RT microRNA PCR (Exiqon, Denmark) kit, and the expression level of U6 snRNA was used as The expression difference of miR-17-5p and miR-20a mature body miRNA in K562 cells and HL60 cells was analyzed by internal control. The results showed that the expression levels of miR-17-5p and miR-20a mature body miRNA in K562 cells were higher than those in HL60 cells, about 3 times that of HL60 cells ( figure 2 A).
[0044] The K562 cells and HL60 cells in the logarithmic growth phase were collected, the total cell protei...
Embodiment 3
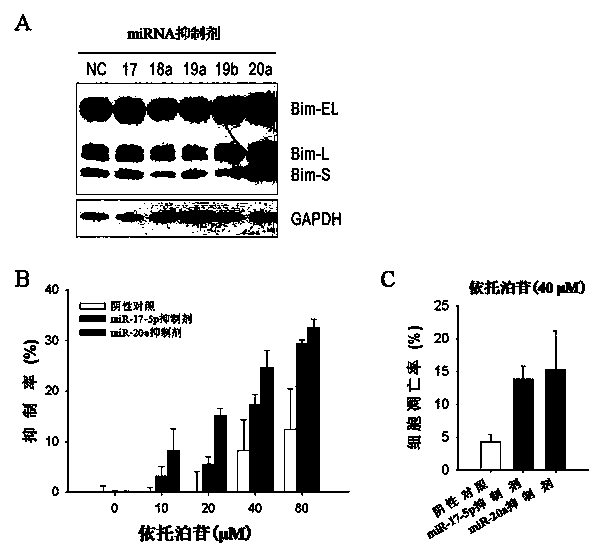
[0045] Example 3 Effect of inhibiting miR-17-5p, miR-20a or overexpressing Bim-S on apoptosis
[0046] To study the role of miR-17-5p and miR-20a in apoptosis, we transfected single-chain miR-17-5p or inhibitor of miR-20a into K562 cells using Neon electrotransfection system. After 48 hours, cells were collected to detect cell apoptosis by AnnexinV-FITC / PI staining. Compared with the negative control, inhibitors of miR-17-5p or miR-20a could significantly increase the proportion of K562 cells undergoing apoptosis ( image 3 A).
[0047] In order to study the effect of Bim-S protein on apoptosis, we constructed an expression vector to overexpress Bim-S. First, use the following primers (synthesized by Guangzhou Yingwei Jieji Co., Ltd.) to amplify the Bim cDNA in K562 cells:
[0048] Upstream primer: TTGGATCCCGCCACCATGGCAAAGCAACCTTCTGATG,
[0049] Downstream primer: TTGAATTCTCAATGCATTCTCCACACCAGG.
[0050] The DNA band whose size conforms to Bim-S was recovered from rubbe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com